I don’t disagree with you for a second that SimpleITK needs to be compatible with ITK. Why in heavens would not be? That wasn’t the spirit of my question.
I am thinking about interoperability between SimpleITK and Numpy.
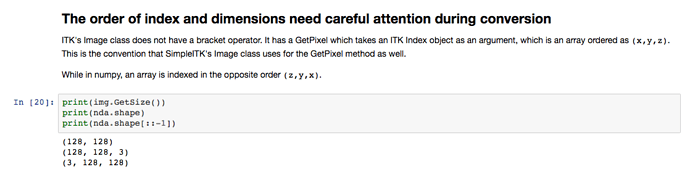
What I am thinking is why do we need to flip the order of the indices when getting the numpy arrays??
Notice that in your example you pass the sitk image to imshow, it makes sense that it works since the internal layout is [x,y,z] as show consistently throughout the sitk image methods. So no problem there.
But there is the expectation operate in the same manner with the numpy array, but you can’t because the indices are reversed.
So if I were going to extract the numpy array and do any kind of operation and then visualize it, you have to be constantly aware of this change or your numpy processing code won’t work or neither the visualization.
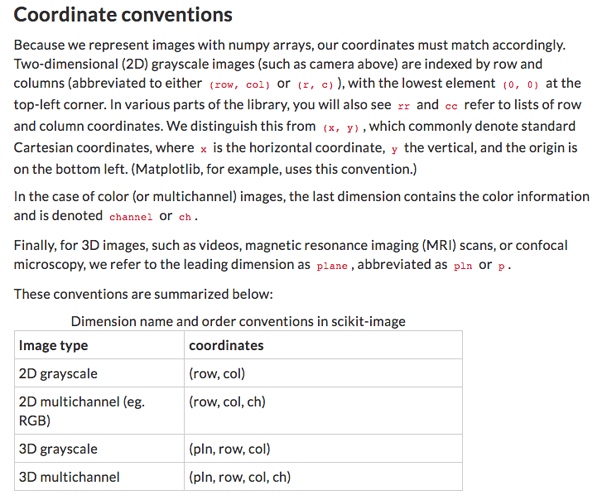
Maybe this [z, y, x] ordering is compatible with numpy as you suggest but so are other orderings like [z,x,y] or [y,z,x] but I would debate that computational efficiency is relative to the way the programmer handles his/her arrays.
Every time I pass from sitk to numpy and back I need to be aware of this change. That reduces compatibility, and creates the perfect situation to introduce bugs in your code.
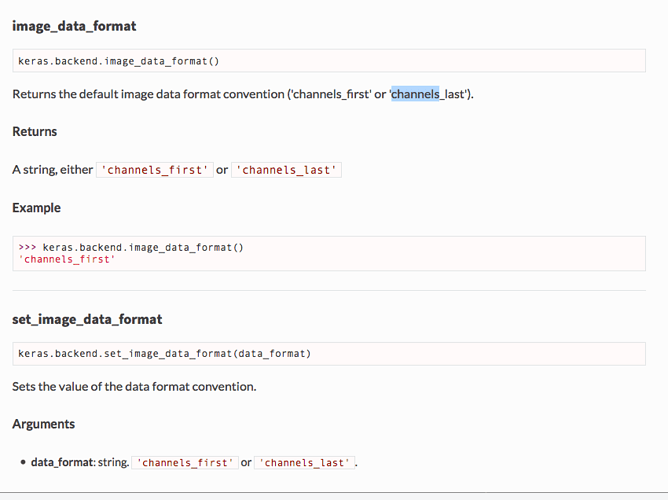
My suggestion is to have an API method that enables you to return numpy arrays with the order that you expect, in a similar fashion to the example I provided earlier for keras.
Something like:
...
sitk.SetArrayLayout(sitk.DEPTH_FIRST)
narr = sitk.GetArrayFromImage(img) # returns array as [z,y,x]
sitk.SetArrayLayout(sitk.DEPTH_LAST)
narr = sitk.GetArrayFromImage(img) # returns array as [x,y,z]