Hi!
I’m building an image registration framework for MRI data that requires combination of numpy and ITK. I recently realised that the indexing in ITK arrays and numpy arrays are swapped from [x,y,z] to [z,y,x] (from this forum thread). So in order to fix this I thought I’d apply numpy.tranpose before creating the image, but it turns out this has no effect on the generated image. I know that it is not recommended to modify the data like this but I want to make so that translations estimated in “X” in ITK corresponds to the 0th dimension of my numpy array (if there is a better version to do this I’m all ears).
To demonstrate the issue with an example: I have a Shepp logan phantom that I have generated using Sigpy here called phan
phan = abs(sigpy.shepp_logan((64,64,64)))
# Transpose array and create ITK image objects
phan_T = np.transpose(phan, (2,1,0))
img_phan = itk.image_from_array(phan)
img_phan_T = itk.image_from_array(phan_T)
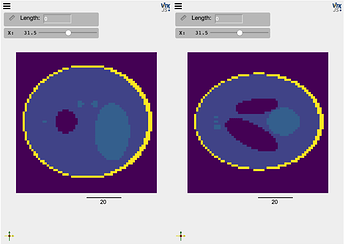
If I compare the numpy arrays I can see the effect of the transpose:
compare(phan, phan_T, interpolation=False, ui_collapsed=True, mode='x')
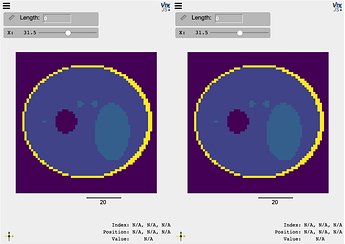
But the effect of the transpose does not appear when I compare the ITK images
compare(img_phan, img_phan_T, interpolation=False, ui_collapsed=True, mode='x')
My thought is that this has something to do with the fact that numpy is returning a view of the array and not a deep copy, but I’ve tried creating a copy of the array before producing the ITK image but with the same result. The only way I could get it to work was to create a 4D array which I slice in the transpose function as
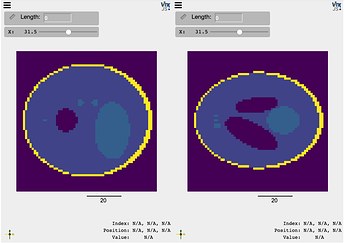
A = np.zeros((64,64,64,2))
A[:,:,:,0] = phan
phan_T = np.transpose(A[:,:,:,0], (2,1,0))
img_phan = itk.image_from_array(phan)
img_phan_T = itk.image_from_array(phan_T)
compare(img_phan, img_phan_T, interpolation=False, ui_collapsed=True, mode='x')
I feel like there is something I’m missing here in the way I’m creating the images, any help is very welcome.
Thank you very much!


 )
)