Hello Everyone,
Can you please help me with the below error?
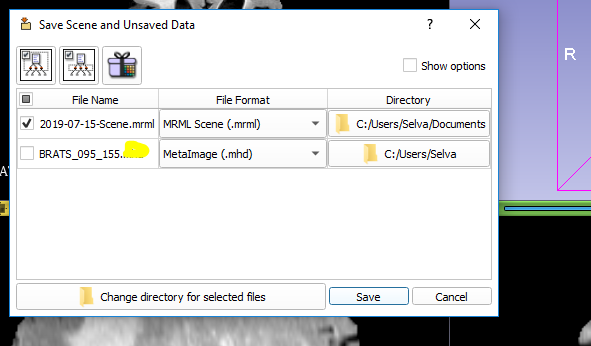
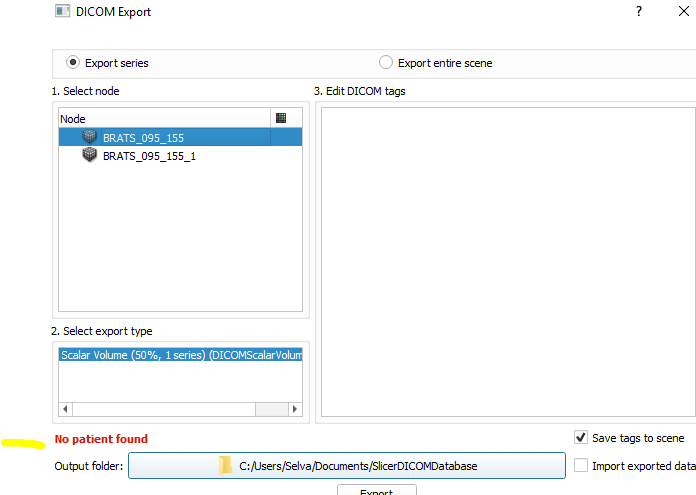
I have a DICOM file which is attached. When we try to convert this into a.mhd file, we get an error as “Teh file in the series has unsupported 3D dimensions”. Can you please help us resolve the issue?
II am also providing the tags information of my file based on code from this link (https://simpleitk.readthedocs.io/en/master/Examples/DicomImagePrintTags/Documentation.html)
(0008|0008) = = “ORIGINAL\PRIMARY\VOLUME\NONE”
(0008|0016) = = “1.2.840.10008.5.1.4.1.1.130”
(0008|0018) = = “2.25.268344867590623757739343791212928671539”
(0008|0020) = = “20190714”
(0008|0023) = = “20190714”
(0008|002a) = = "19700101070000 "
(0008|0030) = = “172248”
(0008|0033) = = “173525”
(0008|0050) = = " "
(0008|0060) = = “PT”
(0008|0070) = = "UNKNOWN "
(0008|0090) = = “”
(0008|1030) = = "UNKNOWN "
(0008|103e) = = "UNKNOWN "
(0008|1090) = = "UNKNOWN "
(0008|9205) = = “MONOCHROME”
(0008|9206) = = “VOLUME”
(0008|9207) = = "VOLUME_RENDER "
(0010|0010) = = “abcd”
(0010|0020) = = “2000”
(0010|0030) = = " "
(0010|0040) = = “Male”
(0018|0050) = = "1 "
(0018|0071) = = “MANU”
(0018|0073) = = “MANU”
(0018|1000) = = "UNKNOWN "
(0018|1020) = = "UNKNOWN "
(0018|1134) = = “STATIC”
(0018|1181) = = “NONE”
(0018|2005) = = "-78-77-76-75-74-73-72-71-70-69-68-67-66-65-64-63-62-61-60-59-58-57-56-55-54-53-52-51-50-49-48-47-46-45-44-43-42-41-40-39-38-37-36-35-34-33-32-31-30-29-28-27-26-25-24-23-22-21-20-19-18-17-16-15-14-13-12-11-10-9-8-7-6-5-4-3-2-1\0\1\2\3\4\5\6\7\8\9\10\11\12\13\14\15\16\17\18\19\20\21\22\23\24\25\26\27\28\29\30\31\32\33\34\35\36\37\38\39\40\41\42\43\44\45\46\47\48\49\50\51\52\53\54\55\56\57\58\59\60\61\62\63\64\65\66\67\68\69\70\71\72\73\74\75\76 "
(0018|9004) = = “RESEARCH”
(0018|9073) = = “0”
(0018|9725) = = “CYLINDRICAL_RING”
(0018|9726) = = “0”
(0018|9727) = = “0”
(0018|9755) = = "FALSE "
(0018|9758) = = “NO”
(0018|9759) = = “NO”
(0018|9760) = = “NO”
(0018|9761) = = “NO”
(0018|9762) = = “NO”
(0018|9763) = = “NO”
(0018|9764) = = “NO”
(0018|9765) = = “NO”
(0018|9766) = = “NO”
(0018|9767) = = “NO”
(0018|9768) = = “NO”
(0020|000d) = = “1.2.826.0.1.3680043.2.1125.4082960453717943603603726631811946355”
(0020|000e) = = “1.2.826.0.1.3680043.2.1125.8397261186222101954520583083004172948”
(0020|0010) = = " "
(0020|0011) = = “”
(0020|0013) = = "0 "
(0020|0032) = = "-120-120-78 "
(0020|0037) = = "0.99999999999999\0\0\0\0.99999999999999\0 "
(0020|0052) = = “2.25.183387019459409372524007875133033107487”
(0020|1040) = = “”
(0028|0002) = = “1”
(0028|0004) = = "MONOCHROME2 "
(0028|0008) = = "155 "
(0028|0010) = = “240”
(0028|0011) = = “240”
(0028|0030) = = "1\1 "
(0028|0100) = = “16”
(0028|0101) = = “16”
(0028|0102) = = “15”
(0028|0103) = = “0”
(0028|0106) = = “0”
(0028|0107) = = “2213”
(0028|0108) = = “0”
(0028|0109) = = “2213”
(0028|0301) = = “NO”
(0028|1050) = = “1106.5”
(0028|1051) = = “2213”
(0028|2110) = = “00”
(0054|0202) = = “STATIONARY”
(0054|1002) = = “EMISSION”
(0054|1210) = = "0 "
(2050|0020) = = “IDENTITY”
Image Size: (240, 240, 155)
Image PixelType: 16-bit unsigned integer