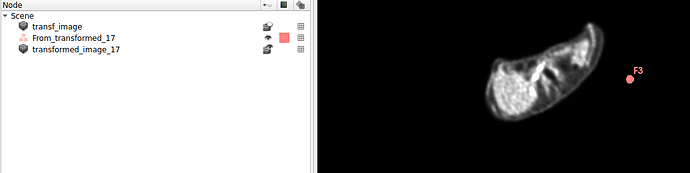
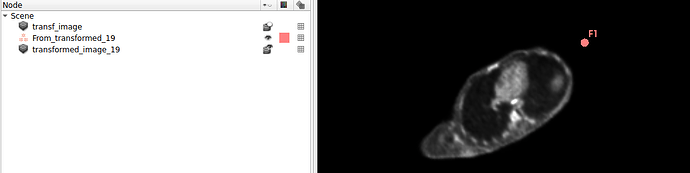
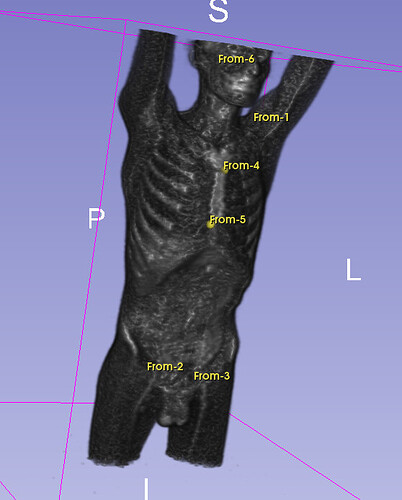
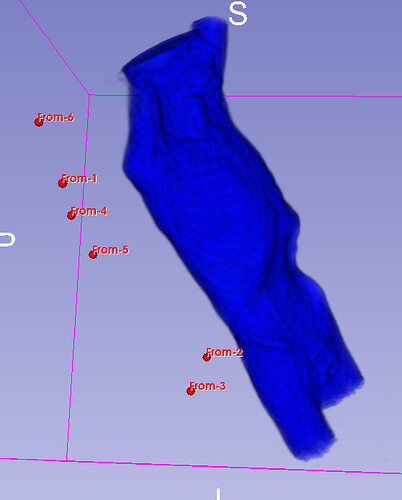
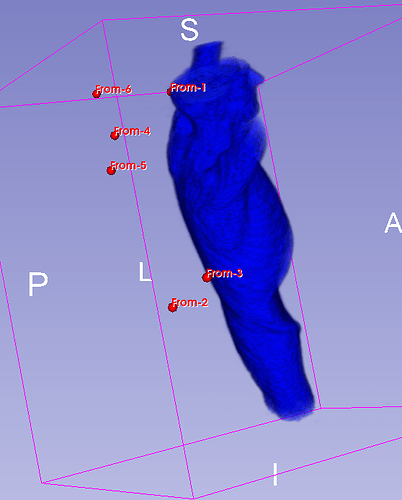
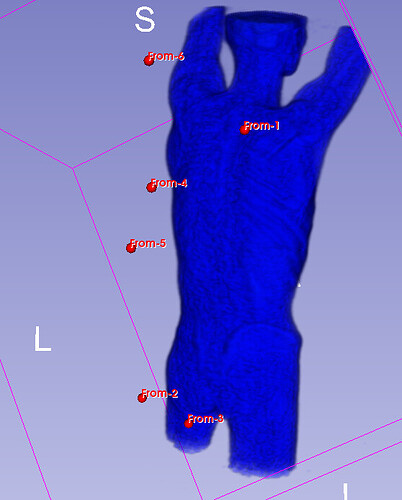
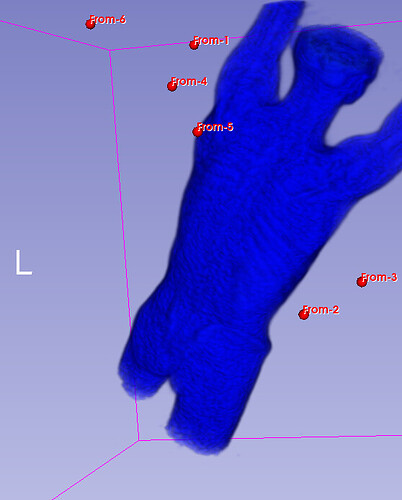
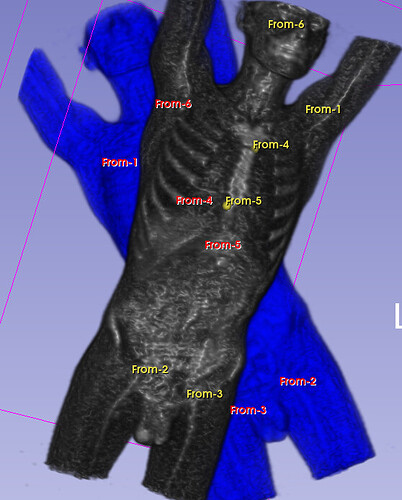
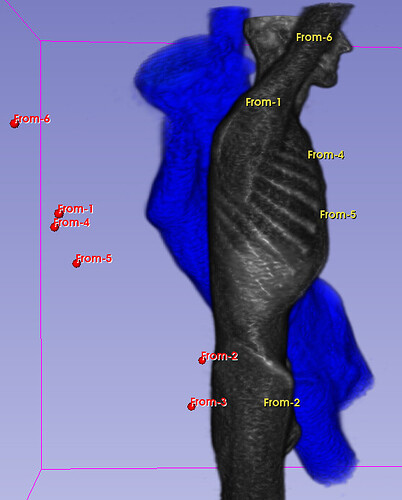
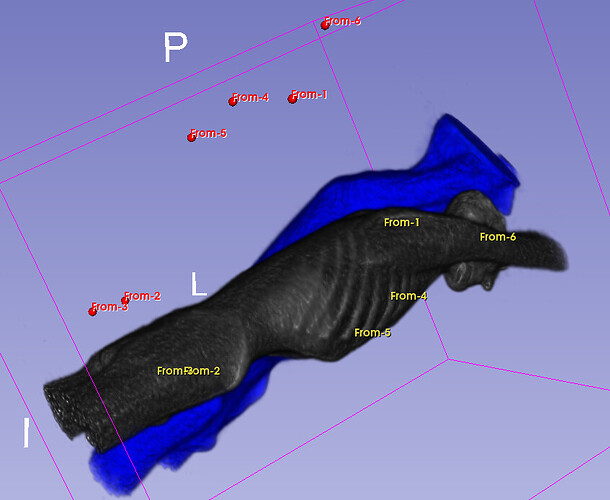
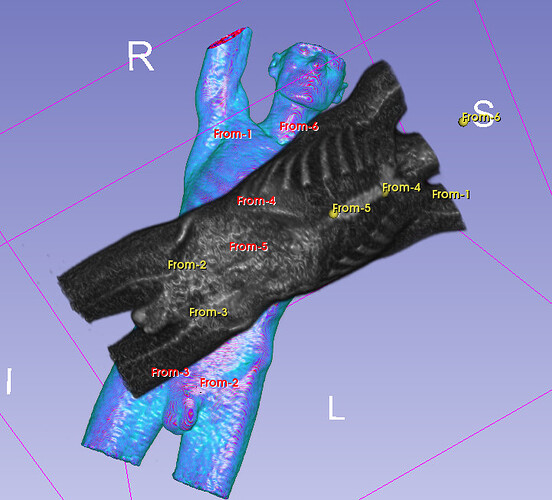
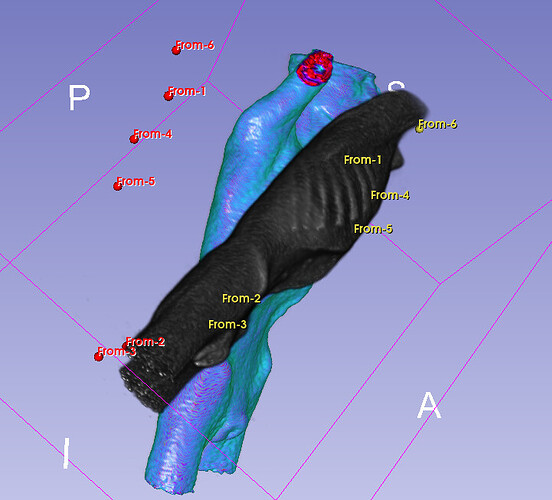
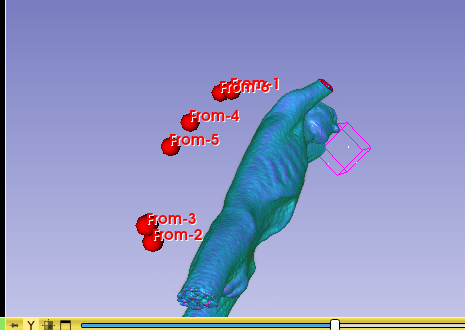
Hello I try to rotate an image and landmarks , around image center. I tried resampling landmarks boh by resample physical point and manually using rotation matrix (I would prefer to stic with second solution as I need to make it differentiable in the end). However it do not work Image get rotated but fiducial points are not staying in the same anatomical positions (they get generally get outside of the body after rotation).
code below, I attach also image and landmark points
import SimpleITK as sitk
import numpy as np
from jax.scipy.spatial.transform import Rotation
def save_landmarks_for_slicer(points, out_file_path,original_json_path):
with open(original_json_path) as f:
data = json.load(f)
for i in range(len(cps)):
data['markups'][0]['controlPoints'][i]['position']=points[i]
with open(out_file_path, 'w') as f:
json.dump(data, f)
landmarks=np.load('/workspaces/pilot_lymphoma/data/pat_3/lin_transf/From.npy')
image=sitk.ReadImage('/workspaces/pilot_lymphoma/data/pat_3/lin_transf/study_0_SUVS.nii.gz')
# Assume 'image' is your image and 'landmarks' is your list of landmark points
# weights=jnp.ones(6)*30
weights=np.ones(6)*30
Rotationn=Rotation.from_euler('xyz', weights[0:3], degrees=True)
quaternion=np.array(Rotationn.as_quat())
# Get the center of the image
center = image.TransformContinuousIndexToPhysicalPoint([(index - 1) / 2.0 for index in image.GetSize()])
# Create a VersorRigid3DTransform
transform = sitk.VersorRigid3DTransform()
transform.SetCenter(center)
transform.SetRotation([float(quaternion[0]),float(quaternion[1]),float(quaternion[2]),float(quaternion[3])])
# Resample the image
resampler = sitk.ResampleImageFilter()
resampler.SetOutputDirection(image.GetDirection())
resampler.SetOutputOrigin(image.GetOrigin())
resampler.SetOutputSpacing(image.GetSpacing())
resampler.SetSize(image.GetSize())
resampler.SetTransform(transform)
resampled_image = resampler.Execute(image)
sitk.WriteImage(resampled_image, "/workspaces/pilot_lymphoma/data/transformed_image.nii.gz")
rotation_matrix =np.array(Rotationn.as_matrix())
resampled_landmarks= (landmarks - center) @ rotation_matrix.T + center
save_landmarks_for_slicer(resampled_landmarks, "/workspaces/pilot_lymphoma/data/From_transformed.mrk.json",'/workspaces/pilot_lymphoma/data/pat_3/lin_transf/From.mrk.json')
From.npy (272 Bytes)
From.mrk.json (5.1 KB)