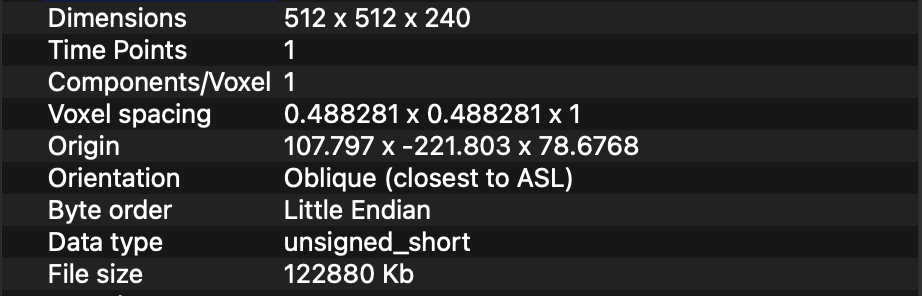
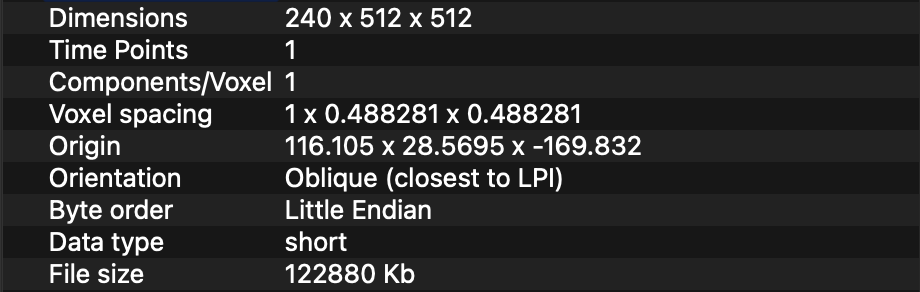
I have a set of 4 DICOM CT Volumes which I am reading with SimpleITK ImageSeriesReader. Two of the images represent the CT of patient before and after the surgery. The other two images are binary segmentation masks segmented on the former 2 CT images. The segmentations are a ROI of their source CT.
All the 4 CT images, have different Size, Spacing, Origin and Direction. I have tried applying this GitHub gist https://gist.github.com/zivy/79d7ee0490faee1156c1277a78e4a4c4 to resize my images to 512x512x512 and Spacing 1x1x1. However, it seems to only work for the source CT images. It doesn’t work on the segmentation images. The segmented structure is always placed in the center of the CT image, instead of the correct location.
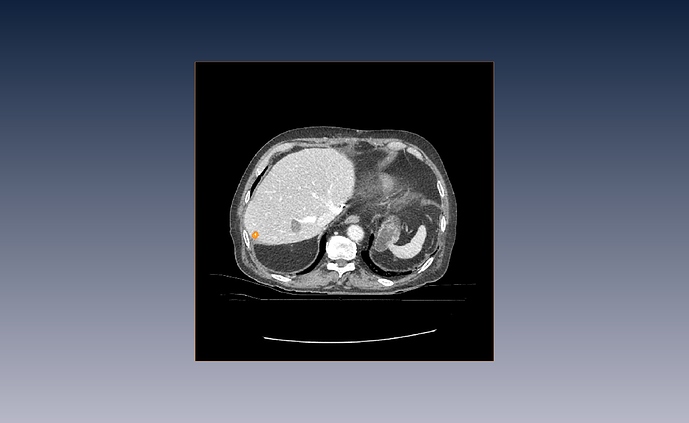
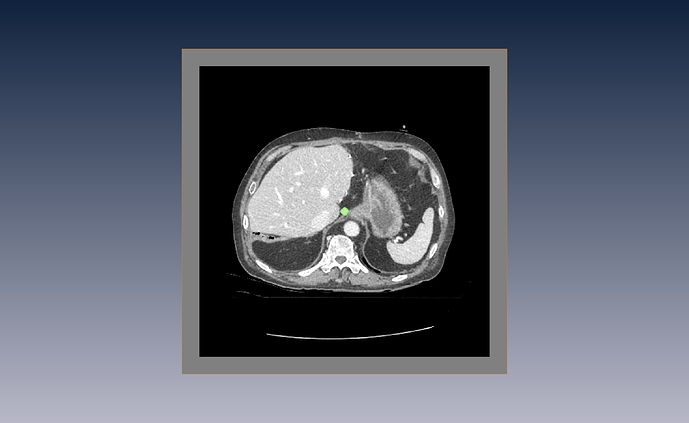
This are my “raw” image with its corresponding segmentation.
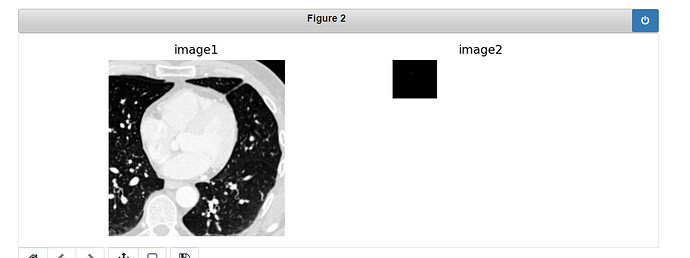
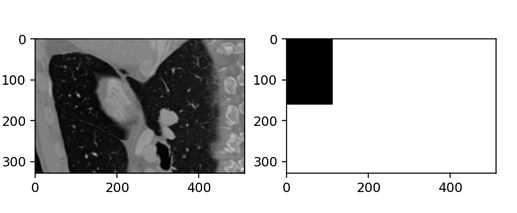
And this is the result which is obviously incorrect.
This is my code:
def resize_resample_images(images):
#Define tuple to store the images
tuple_resized_imgs = collections.namedtuple('tuple_resized_imgs', ['img_plan', 'img_validation', 'ablation_mask', 'tumor_mask'])
# %% Create Reference image with zero origin, identity direction cosine matrix and isotropic dimension
dimension = images.img_plan.GetDimension()
reference_direction = np.identity(dimension).flatten()
reference_size_x = 512
reference_origin = np.zeros(dimension)
data = [images.img_plan, images.img_validation, images.ablation_mask, images.tumor_mask]
reference_physical_size = np.zeros(dimension)
for img in data:
reference_physical_size[:] = [(sz - 1) * spc if sz * spc > mx else mx for sz, spc, mx in
zip(img.GetSize(), img.GetSpacing(), reference_physical_size)]
reference_spacing = [reference_physical_size[0] / (reference_size_x - 1)] * dimension
reference_size = [int(phys_sz/(spc) + 1) for phys_sz,spc in zip(reference_physical_size, reference_spacing)]
reference_image = sitk.Image(reference_size, images.img_plan.GetPixelIDValue())
reference_image.SetOrigin(reference_origin)
reference_image.SetSpacing(reference_spacing)
reference_image.SetDirection(reference_direction)
reference_center = np.array(
reference_image.TransformContinuousIndexToPhysicalPoint(np.array(reference_image.GetSize()) / 2.0))
data_resized = []
for idx,img in enumerate(data):
#%% Set Transformation
transform = sitk.AffineTransform(dimension) # use affine transform with 3 dimensions
transform.SetMatrix(img.GetDirection()) # set the cosine direction matrix
# TODO; set the center correctly
# transform.SetCenter(reference_center)
transform.SetTranslation(np.array(img.GetOrigin()) - reference_origin) # set the translation.
# Modify the transformation to align the centers of the original and reference image instead of their origins.
centering_transform = sitk.TranslationTransform(dimension)
img_center = np.array(img.TransformContinuousIndexToPhysicalPoint(np.array(img.GetSize()) / 2.0))
centering_transform.SetOffset(np.array(transform.GetInverse().TransformPoint(img_center) - reference_center))
centered_transform = sitk.Transform(transform)
centered_transform.AddTransform(centering_transform)
#%% set all output image parameters: origin, spacing, direction, starting index, and size.
resampler = sitk.ResampleImageFilter()
resampler.SetReferenceImage(reference_image)
resampler.SetDefaultPixelValue(img.GetPixelIDValue())
resampler.SetTransform(centered_transform)
resampler.SetSize(reference_image.GetSize())
resampler.SetOutputSpacing(reference_image.GetSpacing())
resampler.SetOutputOrigin(reference_image.GetOrigin())
resampler.SetOutputDirection(reference_image.GetDirection())
if idx==0 or idx==1:
resampler.SetInterpolator(sitk.sitkLinear)
elif idx==2 or idx==3:
resampler.SetInterpolator(sitk.sitkNearestNeighbor)
resampled_img = resampler.Execute(img)
data_resized.append(resampled_img)
resized_imgs = tuple_resized_imgs(img_plan=data_resized[0],
img_validation=data_resized[1],
ablation_mask=data_resized[2],
tumor_mask=data_resized[3])
return resized_imgs