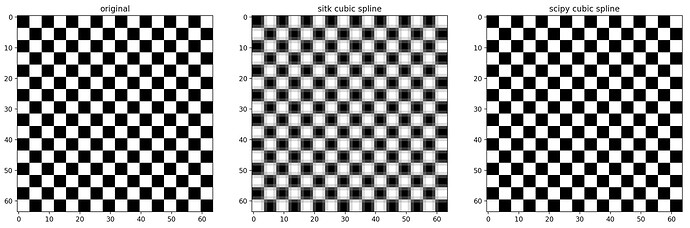
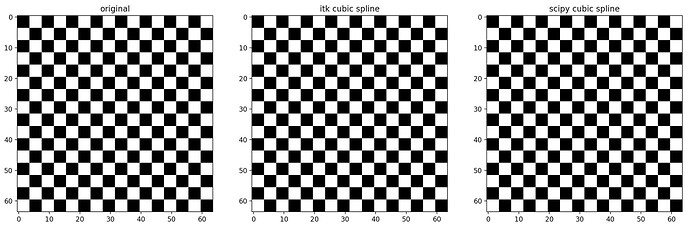
I found out I was getting slightly blurry results when using Resample just with cropping:
import SimpleITK as sitk
import matplotlib.pyplot as plt
import numpy as np
def myshow(img, title=None, margin=0.05, dpi=80):
nda = sitk.GetArrayViewFromImage(img)[:,:,62]
spacing = img.GetSpacing()
ysize = nda.shape[0]
xsize = nda.shape[1]
figsize = ((1 + margin) * ysize / dpi, (1 + margin) * xsize / dpi)
fig = plt.figure(title, figsize=figsize, dpi=dpi)
ax = fig.add_axes([margin, margin, 1 - 2*margin, 1 - 2*margin])
extent = (0, xsize*spacing[1], 0, ysize*spacing[0])
t = ax.imshow(nda,
extent=extent,
interpolation='none',
cmap='gray',
origin='lower',
vmin=0.0, vmax=1.0)
if(title):
plt.title(title)
plt.show()
if __name__ == '__main__':
scale = 1.0
spacing = 0.2
image_size = 250
patch_size = 125
sampling = sitk.sitkBSplineResamplerOrder3
#sampling = sitk.sitkNearestNeighbor
grid = ((np.indices(np.repeat(image_size, 3)) // 4).sum(axis=0) % 2).astype(float)
grid = sitk.GetImageFromArray(grid)
grid.SetSpacing(np.ones(3) * spacing)
grid.SetOrigin((30.9, 10.2, 28.4))
#myshow(grid, 'Grid Input')
outputSpacing = scale * np.array(grid.GetSpacing())
outputDirection = np.diag([1.0, 1.0, 1.0]) @ np.array(grid.GetDirection()).reshape((3,3))
imageCenter = 0.5 * (image_size - 1) * np.array(grid.GetSpacing())
patchCenter = outputDirection.dot(0.5 * (patch_size - 1) * outputSpacing)
translation = imageCenter - patchCenter
print(translation)
outputOrigin = np.array(grid.GetOrigin())# + translation
resampled1 = sitk.Resample(
grid, (np.repeat(patch_size, 3)).tolist(),
sitk.Transform(3, sitk.sitkIdentity), sampling,
defaultPixelValue=255,
outputSpacing=outputSpacing, outputOrigin=outputOrigin, outputDirection=outputDirection.flatten()
)
myshow(resampled1, 'Resampled')
Even if you disable cropping by making patch_size = 250, the result is blurry, as can be seen in the close-up below.
Why is this happening? Is it due to numerical accuracy? Can I alleviate this problem?
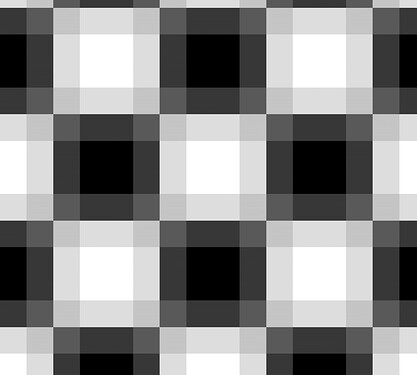
 .
.