Hello itk community,
My goals is to create a code in simple itk to perform quality contrôle of geometri distortion on MRI.
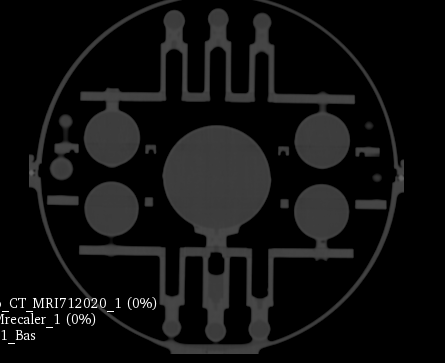
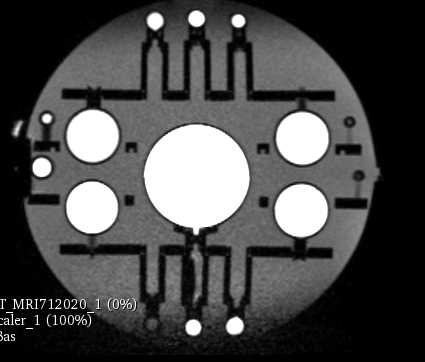
The key for following the evolution of the system performances is to register every new MRI with CT. The CT will be the standard as there is no geometric distortion of it. The image is a mahphan phantom.
CT:0.5x0.5x0.5mm^3
MRI:1x1x1mm^3
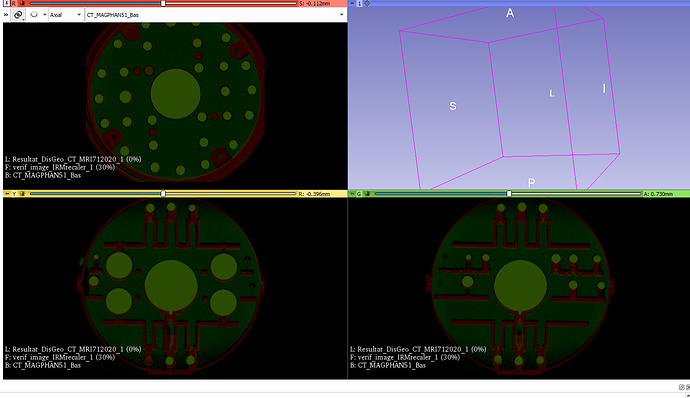
I did not succed to accuratly (less than 1mm) register the two image with the following code.
Can you give me some advice.
def multires_registration(fixed_image, moving_image, initial_transform, ImageSamplingPercentage):
registration_method = sitk.ImageRegistrationMethod()
registration_method.SetMetricAsMattesMutualInformation(numberOfHistogramBins=50)
registration_method.SetMetricSamplingStrategy(registration_method.RANDOM)
registration_method.SetMetricSamplingPercentage(float(ImageSamplingPercentage)/100)
registration_method.SetInterpolator(sitk.sitkLinear)
registration_method.SetOptimizerAsGradientDescent(learningRate=1.0, numberOfIterations=100, estimateLearningRate=registration_method.Once)
registration_method.SetOptimizerScalesFromPhysicalShift()
registration_method.SetInitialTransform(initial_transform)
registration_method.SetShrinkFactorsPerLevel(shrinkFactors = [4,2,1])
registration_method.SetSmoothingSigmasPerLevel(smoothingSigmas = [2,1,0])
registration_method.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
final_transform = registration_method.Execute(fixed_image, moving_image)
print('Final metric value: {0}'.format(registration_method.GetMetricValue()))
print('Optimizer\'s stopping condition, {0}'.format(registration_method.GetOptimizerStopConditionDescription()))
return final_transform
and in the main code:
image_CT = sitk.Cast(image_CT, sitk.sitkFloat64)
image_IRM = sitk.Cast(image_IRM, sitk.sitkFloat64)
initial_transform = sitk.CenteredTransformInitializer(image_CT, image_IRM, sitk.Euler3DTransform(), sitk.CenteredTransformInitializerFilter.MOMENTS)
final_transform =multires_registration(image_CT, image_IRM, initial_transform, 5)
print("main: recalage ok")