Hello, All. I am trying to register vertebra in spine CT and MRI.
The average size of the vertebra MRI is (200-300, 100-200, 15).
The average number of intensity in MRI is around 500. Which is calculated by this.
min_max = np.percentile(new_scan_array.flatten(),[2,98])
num_bins = int(min_max[1]-min_max[0])
The average size of the vertebra CT is (200-300, 100-200, 30-50).
THe average number of intensity in CT is around 1000.
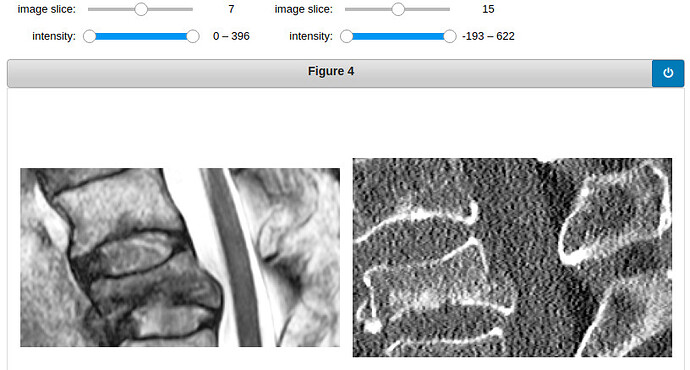
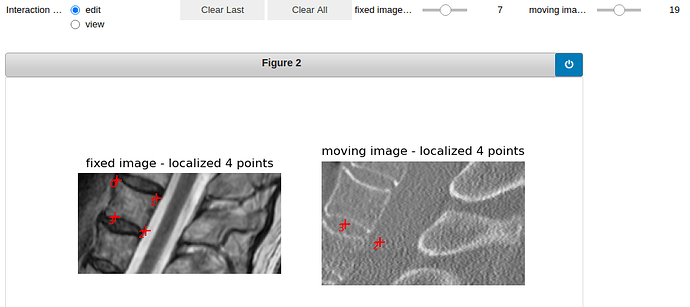
vertebra 19:
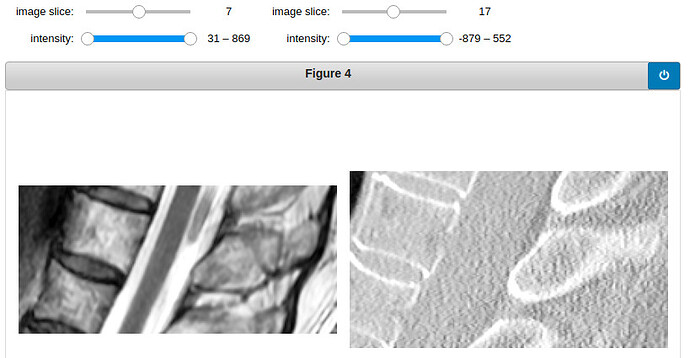
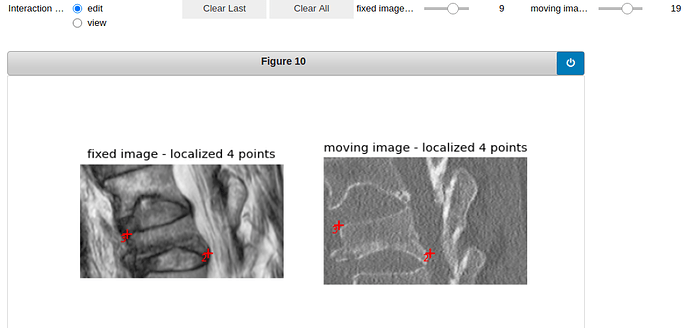
vertebra 9:
Here are two examples of how the crop MRI and CT looks like. (The examples shown are T2-weighted, I have also tried with T1-weighted)
For Initialization, I have used the centorid of the interested vertebra. (The centroids are marked manually in the whole spine scan, and when I cropped the CT and MRI, I maintained the relative position of all vertebrae by resetting the origin.)
initial_transform = sitk.LandmarkBasedTransformInitializer(sitk.VersorRigid3DTransform(),
fixed_image_points_flat,
moving_image_points_flat)
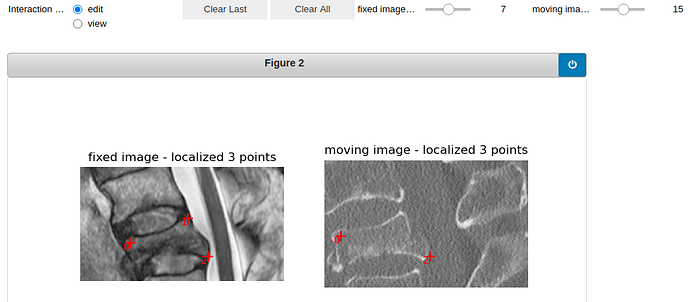
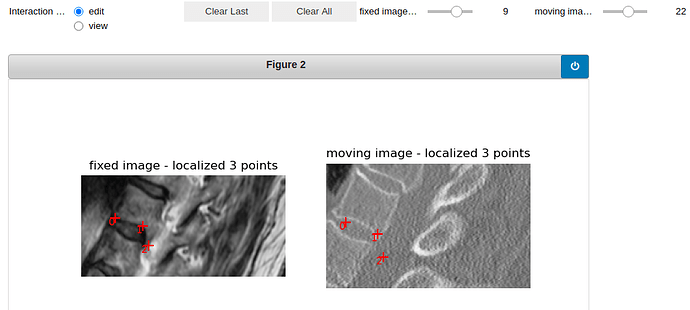
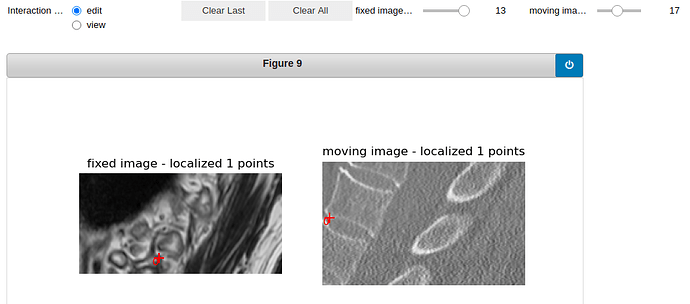
Here is the visualization of the initialization:
vertebra 19:
vertebra 9:
Then I set up the registration method:
registration_method = sitk.ImageRegistrationMethod()
# Similarity metric settings.
registration_method.SetMetricAsMattesMutualInformation(numberOfHistogramBins=1000)
# registration_method.SetMetricAsANTSNeighborhoodCorrelation(radius=20)
registration_method.SetMetricSamplingStrategy(registration_method.NONE)
# registration_method.SetMetricSamplingPercentage(0.1)
registration_method.SetInterpolator(sitk.sitkLinear)
# Optimizer settings.
registration_method.SetOptimizerAsGradientDescent(learningRate=1, numberOfIterations=100,
convergenceMinimumValue=1e-6, convergenceWindowSize=10)
# registration_method.SetOptimizerAsRegularStepGradientDescent(learningRate=1,minStep = 1e-6, relaxationFactor = 0.9,
# numberOfIterations=100)
registration_method.SetOptimizerScalesFromPhysicalShift()
# Setup for the multi-resolution framework.
registration_method.SetShrinkFactorsPerLevel(shrinkFactors = [4,2,1])
registration_method.SetSmoothingSigmasPerLevel(smoothingSigmas=[2,1,0])
registration_method.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
I used MattesMutualInformation and GradientDescent, I set the numberOfHistogramBins to 1000 because the number of intensity values is around 1000. (Please correct me if my intuition is wrong here.)
The numberOfIteration I tired is 100 or 200.
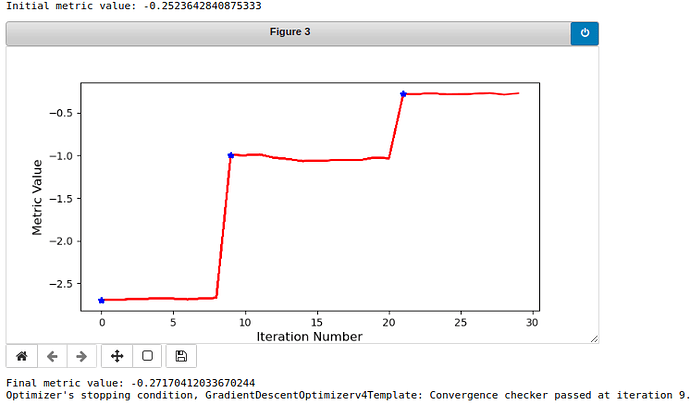
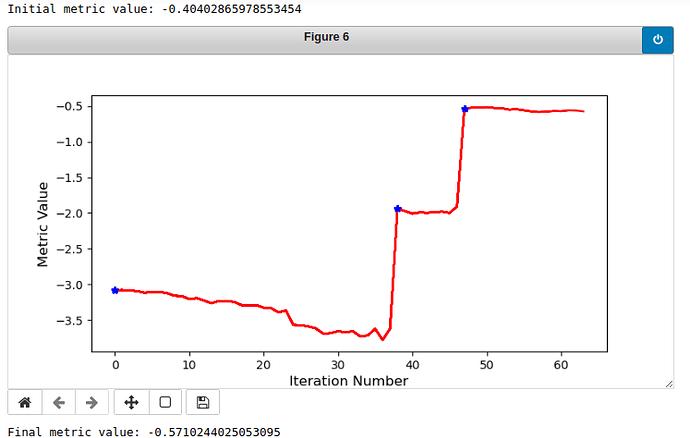
Here are the curves of the metric value:
Vertebra 19:
Vertebra 9:
And here is the final registration result:
Vertebra 19:
Vertebra 9:
As you can see, vertebra 9 is registered well while vertebra 19 is not so good. I want the algorithm to be robust to every vertebra, so do you have any ideas for the method or parameters that I have used? It would be very nice of you to provide some practical advice. If there is more needed to be seen, please inform me.
By the way, I have also tried with the ITK_reigstration_v4 method.
optimized_transform = sitk.VersorRigid3DTransform()
registration_method.SetMovingInitialTransform(initial_transform)
registration_method.SetInitialTransform(optimized_transform, inPlace=False)
final_transform_v4 = registration_method.Execute(fixed_image, moving_image)
final_transform_v4.AddTransform(initial_transform)
But it didn’t work at all, the result made no sense.
Best.