Hi all,
We’re trying to enable 3D Slicer to read and write vector image sequences (displacement field or velocity field: 3D spatial + component + time = 5 dimensions) into a single file. The format of choice is NRRD, and we thought the most correct way would be to use ITK’s NrrdImageIO (the alternative would be to improve vtkTeemNRRDWriter/Reader, but ITK is a much more core dependency than teem).
I started testing the IO with a valid-looking 5D nrrd file that I created using pynrrd. The header of this file contains this:
sizes: 2 20 30 40 4
kinds: time domain domain domain RGBA-color
But when I read it with ITK, and write it back out, the header is changed to:
sizes: 2 20 30 40 1
kinds: time domain domain domain domain
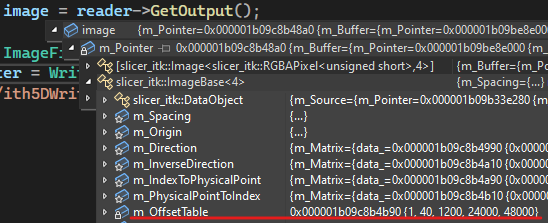
I know that the first issue is at reading, because looking at the offsets, the last dimension is squashed to a single component, see
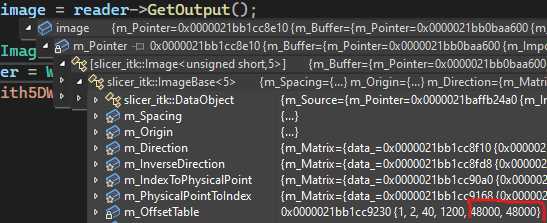
I tried different combinations of “kinds”, but the result was always similar (although the position of the vector component was pushed to the last place:
Original
sizes: 2 4 40 30 20
kinds: time RGBA-color domain domain domain
Write ITK
sizes: 2 40 30 20 1
kinds: time domain domain domain domain
Same thing with
sizes: 4 2 20 30 40
kinds: RGBA-color time domain domain domain
The code I used to read and write the 5D image was this: ITKReadWrite5DImage.cxx · GitHub
My question:
- Is such 5D NRRD IO is supposed to be supported in the first place?
- Is there is something I miss when reading the file? If not, is something broken in ITK?
Any other tips or pointers are appreciated as well. Thank you very much!