Hey!
I have 3-dimensional NIfTI MRI images that I would like to resample to uniform voxel size (1, 1, 1). This works fine:
img = sitk.ReadImage(img_path)
output_spacing = [1.0, 1.0, 1.0]
input_spacing = img.GetSpacing()
input_size = img.GetSize()
size_x = int(round(input_size[0] * input_spacing[0]/output_spacing[0]))
size_y = int(round(input_size[1] * input_spacing[1]/output_spacing[1]))
size_z = int(round(input_size[2] * input_spacing[2]/output_spacing[2]))
size = [size_x, size_y, size_z]
resampler = sitk.ResampleImageFilter()
resampler.SetDefaultPixelValue(0)
resampler.SetOutputDirection(img.GetDirection())
resampler.SetInterpolator(sitk.sitkLinear)
resampler.SetOutputOrigin(img.GetOrigin())
resampler.SetOutputSpacing([1.0, 1.0, 1.0])
resampler.SetSize(size)
img_resampled = resampler.Execute(img)
However, the original images also look rotated:
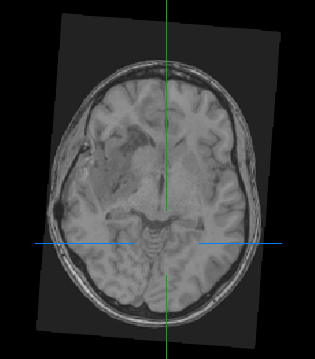
I suspect this is because they have strange directions, e.g. (-0.08117500577885353, -0.01032194044222358, 0. 9966465738475954, 0.9617029292569339, 0.26183942332790555, 0.08103908870110368, 0.26179781186143564, -0.9650562542861514, 0.011325764448564252).
But if I set resampler.SetOutputDirection([1.0, 0.0, 0.0, 1.0, 0.0, 1.0]) instead of resampler.SetOutputOrigin(img.GetOrigin()), the output MRT images have (almost) everywhere the value 0.
What is the reason and how can I fix this error?
Thanks in advance for any help!