I’m trying to extract volume sizes of a liver and tumors from their ground truth volume masks. I’m using the LabelIntensityStatisticsImageFilter, but for one of the inputs it throws me an error:
RuntimeError: Exception thrown in SimpleITK LabelIntensityStatisticsImageFilter_Execute: /tmp/SimpleITK-build/ITK-prefix/include/ITK-5.4/itkImageToImageFilter.hxx:215:
ITK ERROR: LabelImageToStatisticsLabelMapFilter(0x5632f36abe80): Inputs do not occupy the sam e physical space!
InputImage Origin: [-1.0000000e+00, -1.0000000e+00, 1.0000000e+00], InputImage_1 Origin: [-2.4910001e+02, 2.4902318e+02, -6.5100000e+02]
Tolerance: 1.0000000e-06
InputImage Spacing: [1.0000000e+00, 1.0000000e+00, 1.0000000e+00], InputImage_1 Spacing: [9.7656202e-01, 9.7656202e-01, 2.5000000e+00]
Tolerance: 1.0000000e-06
InputImage Direction: -1.0000000e+00 0.0000000e+00 0.0000000e+00
0.0000000e+00 -1.0000000e+00 0.0000000e+00
0.0000000e+00 0.0000000e+00 1.0000000e+00
, InputImage_1 Direction: 1.0000000e+00 0.0000000e+00 0.0000000e+00
0.0000000e+00 -1.0000000e+00 0.0000000e+00
0.0000000e+00 0.0000000e+00 1.0000000e+00
Tolerance: 1.0000000e-06
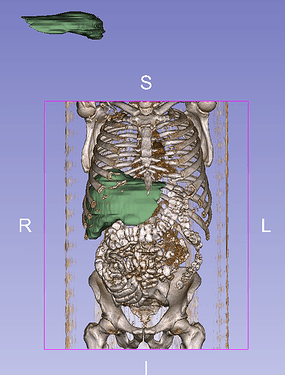
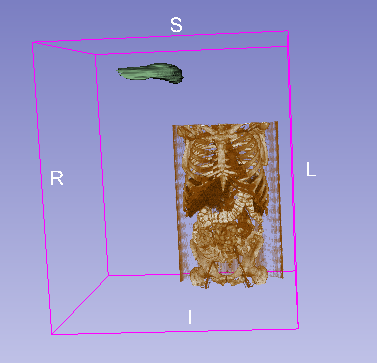
Viewing the inputs confirms that they do not at all occupy the same physical space (see image below), caused by non matching direction, spacing and origin.
I assumed simply resampling the ground truth volume using the intensity volume as reference image would align them (example code below), but that didn’t work.
vol = sitk.ReadImage(vol_path)
gt = sitk.ReadImage(gt_path)
gt_res = sitk.Resample(gt, vol, interpolator=sitk.sitkNearestNeighbor, outputPixelType=gt.GetPixelID())
The resampled volume’s origin, spacing and direction is correct, but it is empty - all voxels have value 0. The same result happens when using any of Slicer’s resample filters. Am I missing something or is this approach straight up wrong?