By: Matt McCormick  , Niels Dekker
, Niels Dekker  , Konstantinos Ntatsis, Marius Staring
, Konstantinos Ntatsis, Marius Staring 
We are exceedingly pleased to announce the release of ITKElastix 0.16.0, an ITK module that provides Python and C++ interfaces to elastix, a toolbox for rigid and nonrigid registration of N-dimensional images. ![]()
![]()
![]()
![]()
Registration is the task of finding a spatial transformation that aligns one image to another by optimizing relevant image similarity metrics.
For more information on how to register scientific images, see the itk-elastix jupyter notebooks and the elastix manual.
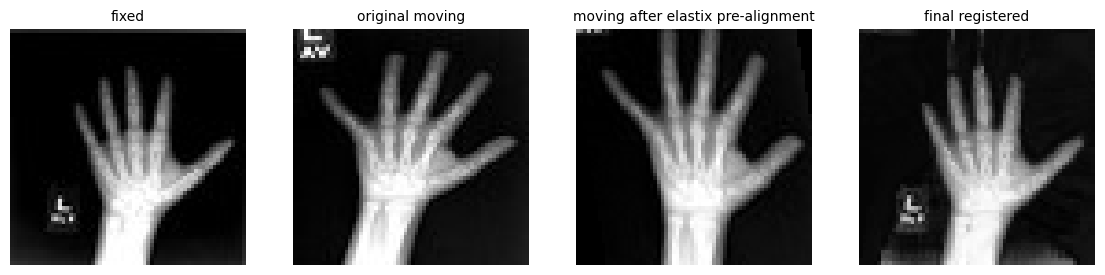
MONAI deep learning registration with elastix pre-alignment (jupyter notebook).
 Download
Download
To install the binary Python packages:
pip install itk-elastix
 Highlights
Highlights
ITKElastix 0.16.0 is released in conjunction with elastix 5.1.0. Features added include:
 Cross-platform binary Python packages are now 80% smaller
Cross-platform binary Python packages are now 80% smaller macOS, Linux ARM Python packages
macOS, Linux ARM Python packages Updated example notebook on MONAI deep learning registration with affine pre-alignment using itk-elastix
Updated example notebook on MONAI deep learning registration with affine pre-alignment using itk-elastix Major performance improvements in the calculation of metrics; accelerates registration for some cases by 40% or more
Major performance improvements in the calculation of metrics; accelerates registration for some cases by 40% or more- C++14 modernization
- Upgraded to ITK 5.3.0
- Support ITK Transform types: Translation, Affine, Euler (2D and 3D), Similarity (2D and 3D), and BSpline
- Add GetCombinationTransform(), GetNumberOfTransforms(), GetNthTransform(n), ConvertToItkTransform(transform) to
itk::ElastixRegistrationMethod - Add SetCombinationTransform(transform) to
itk::TransformixFilter, for access and use of transform objects through the library interface
For more details, see the full elastix 5.1.0 release notes and ITKElastix 0.16.0 release notes.
 What’s Next
What’s Next
In the next release, we will publish additional notebooks on how to support medical image deep learning artificial intelligence (AI) with Project MONAI. These improvement are made in collaboration with the MONAI community as itk Python package spatial metadata improvements in monai harden and extend MONAI’s medical image registration capabilities. We will update the current elastix_napari for napari integration. Also, WebAssembly builds will be created via itk-wasm.
 Acknowledgments
Acknowledgments
ITKElastix was developed in part with support from:
- NIH NIMH BRAIN Initiative under award 1RF1MH126732.
- Chan Zuckerberg Initiative (CZI) Essential Open Source Software for Science award for Open Source Image Registration: The elastix Toolbox.
Enjoy ITK and elastix!