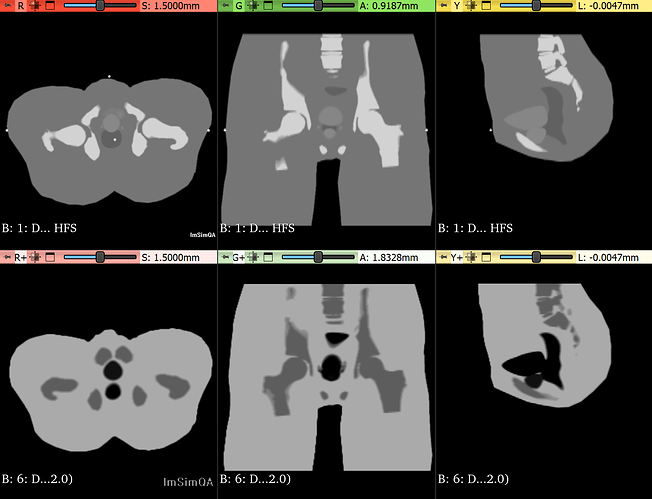
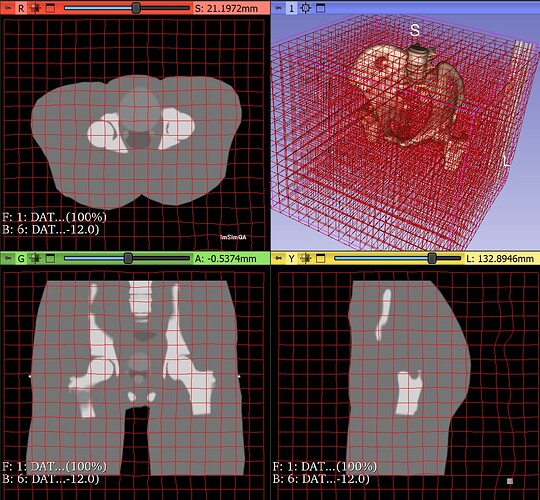
I am trying to register MRI- CT volumes from " Report of the AAPM Radiation Therapy Committee Task Group No. 132" (dataset). The dataset has CT, MR and PET volumes of geometric and anatomical phantoms. The offsets along all axis between the volumes are precisely known. My registration algorithm is converging and results are very close to ground truth but needs some more tuning(Optimizer Regular step gradient descent with Mattes Mutual Information as metric). On close examination I observed that the metric values with ground truth transformation is less than results with my algorithm. It should not be the case. Following example will clarify my problem.
Registration : Phantom Dataset 2 CT volume with Phantom Dataset 1 MRI T2 volume (Rigid registration with translation only) Link
CT Volume size: 512 512 46
CT Volume spacing: 0.7031487 0.7031487 3
CT Volume Direction: 1 0 0 0
CT Volume Origin: -180.23 -180.23 -66.5
MR T2 Volume size: 512 512 46
MR T2 Volume spacing: 0.7031487 0.7031487 3
MR T2 Volume Direction: 1 0 0 0
MR T2 Volume Origin: -180.23 -180.23 -66.5
Ground truth: Offsets in CT, left = 1.0 cm, to anterior = 0.5 cm, to inferior = 1.5 cm
Ground truth Metric Value: 0.8699 (bin size 100)
My registration results:
Center: [-0.575507, -0.575507, 1]
Translation: [-10.2376, 5.28658, -15.0103]
Metric Value: 0.8819 (bin size 100)
Following is how I am estimating the Metric value
Euler3DTransform regTransform = new Euler3DTransform();
regTransform.SetParameters(new VectorDouble() { 0, 0, 0, -10.0, 5.0, -15.0 });
regTransform.SetCenter(new VectorDouble() { -0.575507, -0.575507, 1 });
uint numberOfBins = 100
ImageRegistrationMethod R = new ImageRegistrationMethod();
R.SetMetricAsMattesMutualInformation(numberOfBins);
R.SetMetricSamplingStrategy(ImageRegistrationMethod.MetricSamplingStrategyType.NONE);
R.SetInterpolator(InterpolatorEnum.sitkLinear);
R.SetInitialTransform(regTransform);
double metricValue = R.MetricEvaluate(fixedImage, movingImage);
Even with other metric i.e. Joint Histogram Mutual Information I am facing same issue.
6.2789 (Ground truth) vs 6.3659 (My registration)
Am I missing something here? Could this be the reason registration is not converging towards ground truth?