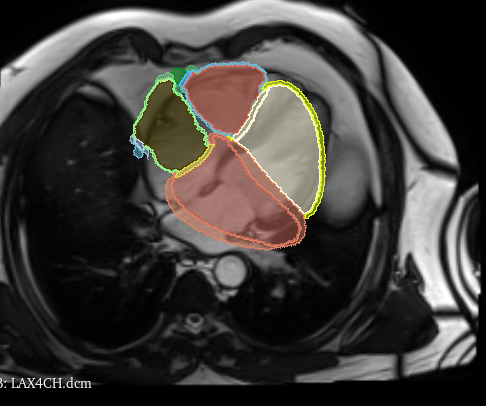
I have a segmentation (original_image) that when I overlap it with the MRI image it comes from it overlaps as expected (comparing it in 3D Slicer, for instance). I perform some operations on Seg3D (or any other software that saves just the image array) and when I save the image (new_image) the overlap doesn’t occur anymore, it’s moved and rotated. I have the function below using SimpleITK (sitk) to try to copy all the possible metadata/header/information from the original_image but it still doesn’t align. The center seems to be fine, but that’s it. I understand that there are probably redundancies in the script, I was trying all possible options.
def save_itk_keeping_header(new_image, original_image, filename):
image_bad_header_itk=sitk.ReadImage(new_image)
image_good_header=sitk.ReadImage(original_image)
image_bad_header_itk.CopyInformation(image_good_header)
image_bad_header_itk.SetDirection(image_good_header.GetDirection())
image_bad_header_itk.SetOrigin(image_good_header.GetOrigin())
[image_bad_header_itk.SetMetaData(key,image_good_header.GetMetaData(key)) for key in image_good_header.GetMetaDataKeys()]
sitk.WriteImage(image_bad_header_itk, filename, True)
The output I get is below - it seems that it should be rotated 90 degrees. Any idea what am I missing? In case it helps, the grey image is an MRI, so they are usually rotated in contrast with CT images that are align with the XYZ coordinate system. Also, I know I’m not copying the spacing - it’s one of the modifications I do - but even if I add it, that doesn’t solve my problem.