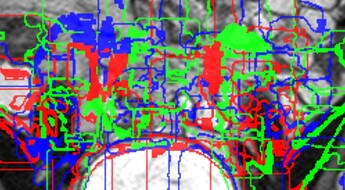
Unfortunately, I have tried all of the pixel types on the list for SimpleITK (see here), and none of them work. I get the same error:
RuntimeError: Exception thrown in SimpleITK LabelMapContourOverlayImageFilter_Execute: /tmp/SimpleITK/Code/Common/include/sitkDualMemberFunctionFactory.hxx:193:
sitk::ERROR: Pixel type: [the pixel type] is not supported in 3D byN3itk6simple33LabelMapContourOverlayImageFilterE
What could I be doing wrong? My full code is simply, for sitkLabelUInt64, for example:
image = sitk.ReadImage("3D_T2_MRI_scan.nii.gz")
slic = sitk.SLICImageFilter()
seg = slic.Execute(denoised_img)
image = sitk.Cast(image, sitk.sitkLabelUInt64)
seg = sitk.Cast(seg, sitk.sitkLabelUInt64)
overlay_filter = sitk.LabelMapContourOverlayImageFilter()
overlay = overlay_filter.Execute(seg, image)
The properties for image are (before casting):
Image (0x18f50190)
RTTI typeinfo: itk::Image<short, 3u>
Reference Count: 1
Modified Time: 12110844
Debug: Off
Object Name:
Observers:
none
Source: (none)
Source output name: (none)
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 12110821
UpdateMTime: 12110843
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [250, 138, 30]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [250, 138, 30]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [250, 138, 30]
Spacing: [0.791016, 0.791016, 3.85]
Origin: [-85.1901, -66.4408, 31.3689]
Direction:
0.99998 5.66689e-06 0.00628384
-0.00628174 -0.0249818 0.999668
0.000162647 -0.999688 -0.0249813
IndexToPointMatrix:
0.791 4.4826e-06 0.0241928
-0.00496895 -0.019761 3.84872
0.000128656 -0.790769 -0.0961781
PointToIndexMatrix:
1.26417 -0.00794136 0.000205618
7.16407e-06 -0.031582 -1.2638
0.00163217 0.259654 -0.00648865
Inverse Direction:
0.99998 -0.00628174 0.000162647
5.66689e-06 -0.0249818 -0.999688
0.00628384 0.999668 -0.0249813
PixelContainer:
ImportImageContainer (0xa468a20)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, short>
Reference Count: 1
Modified Time: 12110840
Debug: Off
Object Name:
Observers:
none
Pointer: 0x2059ebb0
Container manages memory: true
Size: 1035000
Capacity: 1035000
And for seg (before casting):
Image (0xbb15780)
RTTI typeinfo: itk::Image<unsigned int, 3u>
Reference Count: 2
Modified Time: 12111150
Debug: Off
Object Name:
Observers:
none
Source: (0x6010380)
Source output name: Primary
Release Data: Off
Data Released: False
Global Release Data: Off
PipelineMTime: 12111083
UpdateMTime: 12111151
RealTimeStamp: 0 seconds
LargestPossibleRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [250, 138, 30]
BufferedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [250, 138, 30]
RequestedRegion:
Dimension: 3
Index: [0, 0, 0]
Size: [250, 138, 30]
Spacing: [0.791016, 0.791016, 3.85]
Origin: [-85.1901, -66.4408, 31.3689]
Direction:
0.99998 5.66689e-06 0.00628384
-0.00628174 -0.0249818 0.999668
0.000162647 -0.999688 -0.0249813
IndexToPointMatrix:
0.791 4.4826e-06 0.0241928
-0.00496895 -0.019761 3.84872
0.000128656 -0.790769 -0.0961781
PointToIndexMatrix:
1.26417 -0.00794136 0.000205618
7.16407e-06 -0.031582 -1.2638
0.00163217 0.259654 -0.00648865
Inverse Direction:
0.99998 -0.00628174 0.000162647
5.66689e-06 -0.0249818 -0.999688
0.00628384 0.999668 -0.0249813
PixelContainer:
ImportImageContainer (0x17114ea0)
RTTI typeinfo: itk::ImportImageContainer<unsigned long, unsigned int>
Reference Count: 1
Modified Time: 12111092
Debug: Off
Object Name:
Observers:
none
Pointer: 0xd5164f0
Container manages memory: true
Size: 1035000
Capacity: 1035000
I am very confused. The image I’m using is simply a cropped 3D T2 MRI file. It is not corrupted, as I am able to open and view it clearly, and apply filters on it.