Hi all,
I’m working on a workflow that includes image registration, with allowance for rigid, affine or bspline transforms, and initialization using either moments, geometry or landmarks.
Some background:
My workflow was developing quite nicely in SimpleElastix (Selx). In the early days I managed to get from 0 to “something workable” in Selx but I struggled in Sitk. But I eventually ran into some roadblocks with Selx (e.g. bspline registration with landmarks for initial alignment), so I decided to make a move to Sitk.
The data:
DICOM MR images (T1 and T2).
Variability test:
Perform an affine registration using center-of-mass or geometrical center 10 times. For Sitk, also vary the sampling percentage (5%, 10%, 25%, 50%, 75%, 100%) for evaluating the metric. Each “test” consists of 10 repeated runs with a fixed initialization method and sampling percentage (for Sitk).
Gauge performance by performing image addition and subtraction of a single slice in fixed image and registered image (middle of the stack, same slice index for both images), or adding the (10) registered images (pixel-by-pixel, frame-by-frame) from any given test.
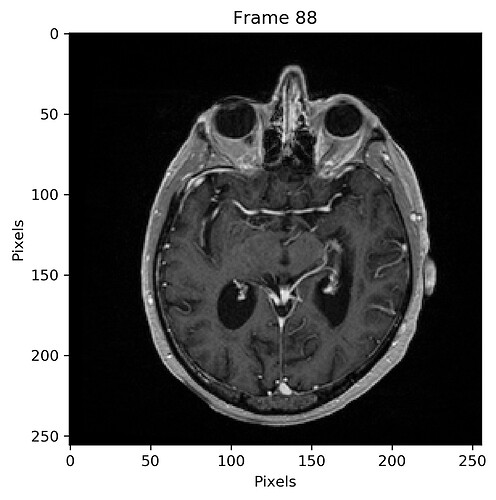
Result using Selx:
Registration takes 20-30 s per run using either COM or geometry. COM and geometry results look different, but results look identical visually when looking at any given initialization method (i.e. COM or geometry).
Following is a plot of the sum of all registered images using a geometrical centering for initialization:
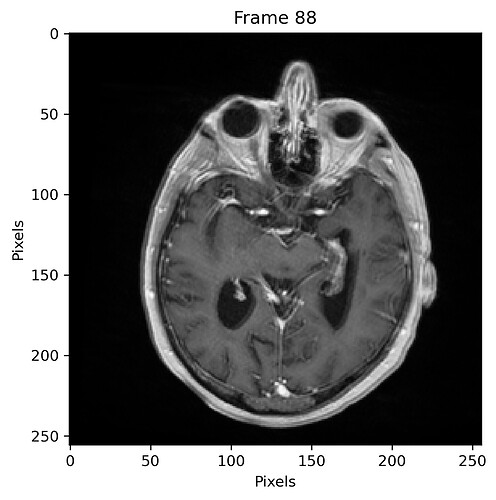
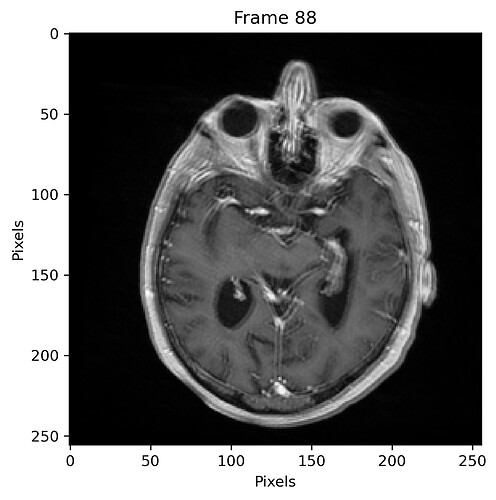
Result using Sitk:
Registration takes 25-80 s per run depending on sampling percentage, using either COM or geometry. Results are highly variable when looking at any given initialization method with any given sampling percentage.
The final metric values are similar from run to run, for different sampling and different initializations, with a similar number of iterations to reach convergence. The registration always terminates due to convergence (as opposed to hitting a limit on the maximum number of iterations).
Following is a plot of the sum of all registered images using a geometrical centering for initialization, and with 5% sampling percentage:
and with 50% sampling percentage:
I estimate the maximum pixel shift to be approximately 3 pix (3 mm).
Sitk Registration method:
def SitkNonDefReg(FixIm, MovIm, Transform='affine', InitMethod='moments',
SamplingPercentage=5, NumIters=500):
import SimpleITK as sitk
from ipywidgets import interact, fixed
def command_iteration(method):
print(f"{method.GetOptimizerIteration():3} = {method.GetMetricValue():10.5f}")
print("\t#: ", len(method.GetOptimizerPosition()))
def command_multi_iteration(method):
print("--------- Resolution Changing ---------")
if InitMethod in ['geometry', 'geometricalcenter']:
CTIF = sitk.CenteredTransformInitializerFilter.GEOMETRY
elif InitMethod in ['moments', 'centerofgravity']:
CTIF = sitk.CenteredTransformInitializerFilter.MOMENTS
SamplingPercentage = SamplingPercentage/100
LearningRate = 1.0
#LearningRate = 5.0
if Transform == 'rigid':
SitkTx = sitk.Euler3DTransform()
elif Transform == 'affine':
SitkTx = sitk.AffineTransform(FixIm.GetDimension())
""" Initial alignment. """
InitialTx = sitk.CenteredTransformInitializer(FixIm, MovIm,
SitkTx, CTIF)
""" Registration. """
RegMethod = sitk.ImageRegistrationMethod()
RegMethod.SetInitialTransform(InitialTx, inPlace=False)
""" Similarity metric settings. """
RegMethod.SetMetricAsMattesMutualInformation(numberOfHistogramBins=50)
RegMethod.SetMetricSamplingStrategy(RegMethod.RANDOM)
RegMethod.SetMetricSamplingPercentage(SamplingPercentage)
""" Setup for the multi-resolution framework. """
ShrinkFactors = [4,2,1]
SmoothingSigmas = [2,1,1]
RegMethod.SetShrinkFactorsPerLevel(shrinkFactors=ShrinkFactors)
RegMethod.SetSmoothingSigmasPerLevel(smoothingSigmas=SmoothingSigmas)
RegMethod.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
""" Interpolator settings. """
RegMethod.SetInterpolator(sitk.sitkLinear)
""" Optimizer settings. """
RegMethod.SetOptimizerAsGradientDescent(learningRate=LearningRate,
numberOfIterations=NumIters,
estimateLearningRate=RegMethod.Once)
RegMethod.SetOptimizerScalesFromPhysicalShift()
# Connect all of the observers for plotting during registration:
RegMethod.AddCommand(sitk.sitkStartEvent, StartPlot)
RegMethod.AddCommand(sitk.sitkEndEvent, EndPlot)
RegMethod.AddCommand(sitk.sitkMultiResolutionIterationEvent, UpdateMultiresIterations)
RegMethod.AddCommand(sitk.sitkIterationEvent, lambda: PlotValues(RegMethod))
#FinalTx = RegMethod.Execute(sitk.Cast(FixIm, sitk.sitkFloat32),
# sitk.Cast(MovIm, sitk.sitkFloat32))
FinalTx = RegMethod.Execute(FixIm, MovIm)
""" Post registration analysis. """
print("-------")
print('InitialTx:')
print(InitialTx)
print("-------")
print("-------")
print('FinalTx:')
print(FinalTx)
print("-------")
print(f'Final metric value: {RegMethod.GetMetricValue()}')
print("Optimizer stopping condition:",
f"{RegMethod.GetOptimizerStopConditionDescription()}")
#print(f"Final Iteration: {RegMethod.GetOptimizerIteration()}")
RegIm = sitk.Resample(MovIm, FixIm, FinalTx, sitk.sitkLinear, 0.0,
MovIm.GetPixelID())
return RegIm
I would very greatful if someone could check over the above method and suggest any ideas on where I’m going wrong, what could be causing the large variability and how to overcome it.
 Many times I’ve gone in circular arguments when trying to decide whether to progress with SimpleElastix or SimpleITK.
Many times I’ve gone in circular arguments when trying to decide whether to progress with SimpleElastix or SimpleITK.

