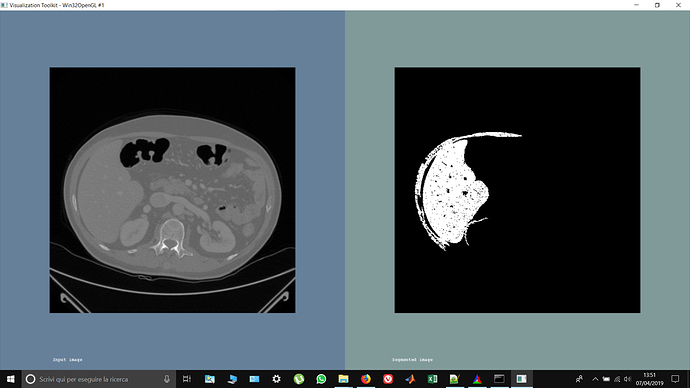
Hi guys, I’ve roughly segmented the liver using confidence connected. I have obtained a binary mask but what I need is to create an image containing ONLY the segmented liver using that mask. What I have to do is like ‘multiplying’ my mask with the image in order to get only the segmented part (the white ones)…how do i do this? I’ve tried doing: image * mask but it doesnt work…
Any help would be much appreciated! Thanks in advance
Hi matt and thank u for your answer. That is exactly what I was looking for, thank u very much!! And thank u again for the ‘‘itkSoftwareGuide’’, it is saving me so much!
Best regards
Hi @matt.mccormick and sorry to bother u! I’ve got this problem… why, in the segmented image, is it gray in the background instead of black?
I think it’s a problem in the parameter i set using confidence connected (for the liver segmentation) and the MaskImageFilter
confidenceConnected->SetMultiplier( 2.2 );
confidenceConnected->SetNumberOfIterations( 5 );
confidenceConnected->SetReplaceValue( 255 );
confidenceConnected->SetSeed( index );
confidenceConnected->SetInitialNeighborhoodRadius( 2 );
voting->SetRadius( indexRadius );
voting->SetBackgroundValue( 0 );
voting->SetForegroundValue( 255 );
voting->SetMajorityThreshold( 2 );
voting->SetMaximumNumberOfIterations( 15 );
Note: before segmentation, I changed the pixel type of the original image from ‘signed short’ to ‘float’ and I rescaled it between 0 and 1… is this actually affecting the MaskImageFilter? One last thing… the intensity of the pixel in the ‘segmented image’ look different from the ‘Input image’, why? Should I care? Should I not perform rescaling?
Thanks in advance for your precious help!
Hi @Marco_Festugato,
Good questions.
We usually just rescale outputs to make it easier for visualization tools. Many visualization tools just support unsigned char pixel types, which means a range of values from 0 to 255, and they do not support dynamically adjusting the window / level, i.e. how values are mapped to colors.
The color black is dependent on the color mapping settings of the visualization tool.
Many tools we work with for image analysis support multiple pixel types and dynamically adjusting the window / level.
Try:
to name a few.
“Black” in CT is -1000, not zero. Zero is water, which appears gray. You should set -1000 as OutsideValue in MaskImageFilter.
Ty again @matt.mccormick for your kind answer. I dont think it’s a visualization problem because I’ve opened one of my images with MATLAB and i verified that i dont have the value of 0 in the background but 1024 
Maybe i should have been more precisely from the beginning…i have 199 dicom images (CT), from which i segmented the liver (but first i converted the pixel type from ‘signed short’ to ‘float’ and rescaled it between 0 and 1). Now it is mandatory for my project to save the 3d volume of only the segmented liver in 199 dicom images… i symply use a CastFilter from ‘float’ to ‘signed short’ and use the example i found in "DicomSeriesReadSeriesWrite.cxx’’ to first read the series in a volume and then write the volume of the segmented liver in a serie of images… should i rescale first the images in the original range before re-casting? Does this anything to do with the fact that i dont have ‘0’ in the background as i want??
Thank u very much for your help!
hi @dzenanz and thank u for your answer. My range is between 0 and 1, so i set the background value to 0… i tried change it but what i got was a binary image 
Marco et al,
I am also looking at segmenting the liver from CT images.
I am interested in automatic techniques.
My best leads are:
- SimpleITK – for loading and preprocessing
- https://github.com/IBBM/Cascaded-FCN – this seems really hopeful
-
https://imagej.net/Trainable_Weka_Segmentation – need Weka models or create them myself
I would be willing to share my best approach with you.
I had the following questions on your approach:
- Are you performing any preprocessing of your CT images to remove noise?
- Is your approach manual, semi-automatic, or fully automatic?
- How did you set your seed points?
- Can you share your ITK prototype source code?
thx in advance
e.-
Hi @keesh!
- Im thinking about performing some noise removal (mean filter) but i havent used it yet;
- semi-automatic, I used a simple region growing technique. It’s not perfect but it works fine for our project;
- using 3D Slicer (from Kitware as well, and free!). U can also use MATLAB;
- Not yet!
Marco,
Thanks for your quick response.
If I achieve decent results on any single stage then I will share them.
thx e.-
If casting and the values are in the range of 0 and 1, most of the information will be lost. 0.1, 0.114, 0.225, etc. all become zero.
Yes. itk::RescaleImageFilter can achieve this.
Marco,
How is your approach going?
Are you ready to share your ITK prototype?
Mine is here-- https://github.com/keesh0/cfcn_test_inference
It works well, but needs a 32 GB GPU + ~40 GB CPU.
Inference is very fast and pretty decent.
thx e.-
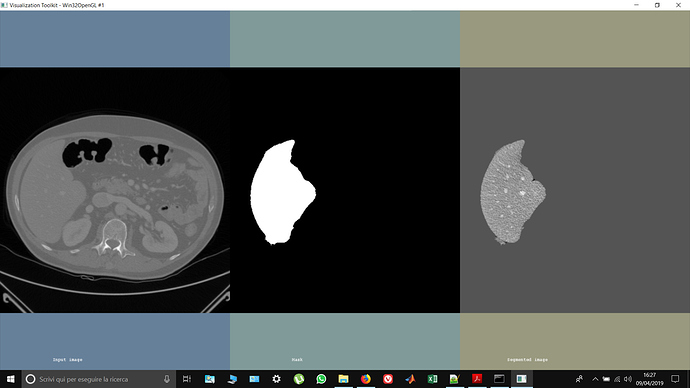
Hi @Marco_Festugato, I see in the image you were able to get rid / clear up the border tissue next to liver segmented area in comparison to the first image you posted. Were you able to do so just by using mask or multiply filter? I am facing similar issue, where I have identified the organ of my interest but need to eliminate surrounding tissue areas that came as output of my thresholding segmentation algorithm. Thanks, Jiten