Hi @zivy,
Unfortunately there still seems to be a small offset, its very confusing.
This is the transform I’m working with:
AffineTransform (0x7fe6f09b0170)
Matrix:
0.999653 0.0144549 0.0220433
-0.0155293 0.99866 0.049376
-0.0213 -0.0497012 0.998537
Offset: [3.55111, 5.85214, -5.28451]
Center: [-5.10695, 11.6622, 19.4729]
Translation: [4.15071, 6.87731, -5.78384]
Inverse:
0.999653 -0.0155293 -0.0213
0.0144549 0.99866 -0.0497012
0.0220433 0.049376 0.998537
Singular: 0
I’ve run the following steps to derive the overall transform T(x):
```overall = np.eye(4)
A = np.array(trans.GetParameters()[:9]).reshape(3,3)
c = np.array(trans.GetFixedParameters())
t = np.array(trans.GetParameters()[9:])
overall[:3,:3] = A
overall[:3,3] = (-A @ c) + t + c ```
Which gives me the following matrix:
array([[ 0.99965251, 0.01445486, 0.02204332, 3.55111468],
[-0.01552929, 0.99865953, 0.049376 , 5.85214032],
[-0.02130005, -0.04970116, 0.99853698, -5.28450612],
[ 0. , 0. , 0. , 1. ]])
Note that the final column does indeed match up to the offset for the transform, so there’s a sanity check on the maths there.
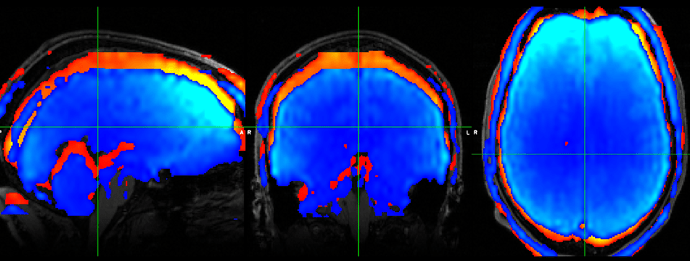
The confusing thing is that there is still some offset between using this matrix and the ground truth I’m working with (an ANTs registered image). The ground truth is in red, the result of using my matrix is in blue, and there is clearly a shift in -z (and also smaller ones in x,y).
Any idea whats happening? It could well be that ANTs is doing something funny internally, and not using all of the parts of the transform (which I realise is an ANTs question, but so far they have been super unhelpful and told me this was an ITK question…)
Tom