I’m trying to detect certain type of artifacts in binary volumes I’m working with, namely step-like transitions between 0 and 1, but only in the three orthogonal planes XY/XZ/YZ. Example:
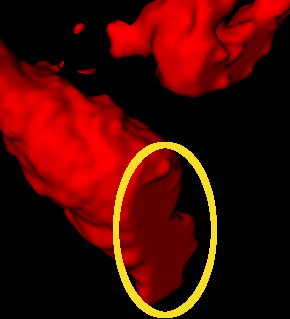
The surface is generally rough and doesn’t end sharply, but in this case it’s truncated in Z direction (the volume is rotated in the screenshot).
What ITK filters can I use to do this efficiently? I don’t need to segment those edges, I just need to know whether they exist in the volume. I would also need to somehow control the size threshold: it’s fine if a 2x2 voxel area vanishes abruplty, but a 20x20 step is something I want to know about.
A simple idea I tried is to convolve an edge-detecting kernel and threshold the output’s absolute intensity (this one is for detecting edges in Z direction only):
kernel_np = np.zeros((2, 20, 20), dtype=np.int16)
kernel_np[0, :, :] = -1
kernel_np[1, :, :] = 1
kernel_sitk = sitk.GetImageFromArray(kernel_np)
output = sitk.Convolution(input, kernel_sitk)
output = sitk.Abs(output)
output = sitk.IntensityWindowing(output, 200.0)
I think I can get it to work with some fine tuning, but it’s quite slow. The binary volumes I’m analysing are of the order of 500x500x500.