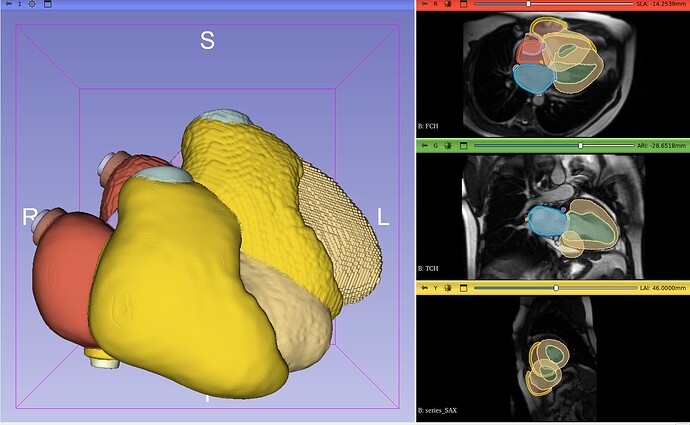
I have a coarse segmentation that I smooth, and the smoothing techniques overwrites the header. I want to retrieve the original one but I have had no success so far. The headers are
_, coarse_header = nrrd.read(path_to_coarse_image)
smooth_image, smooth_header = nrrd.read(path_to_smooth_image)
coarse_header:
OrderedDict([('type', 'short'),
('dimension', 3),
('space', 'left-posterior-superior'),
('sizes', array([748, 749, 749])),
('space directions',
array([[ 0.26375667, -0.09178596, -0.1095799 ],
[ 0.10153998, 0.2821788 , 0.00804705],
[ 0.10060839, -0.04416401, 0.27915496]])),
('kinds', ['domain', 'domain', 'domain']),
('endian', 'little'),
('encoding', 'raw'),
('space origin',
array([-148.47837839, -45.81777566, -68.16309759]))])
smooth_header:
OrderedDict([('type', 'uint8'),
('dimension', 3),
('sizes', array([1314, 884, 900])),
('encoding', 'raw'),
('spacings', array([0.15, 0.15, 0.15])),
('axis mins',
array([-139.14450073, -3.77869463, -26.14629364]))])
Now I don’t care about the information in the smooth_header. The main difference is that it has a higher resolution now, from a voxel spacing of 0.3 now is 0.15.
I figured that the spacing information of the coarse_header is encoded in the norm of the vectors of space_directions, so I copied the the header as follows:
new_smooth_header = dict(coarse_header)
#Update the fields in the new header to match the coarse header, except for the spacing
new_smooth_header['sizes'] = smooth_header['sizes']
smooth_spacing = np.array(smooth_header['spacings']) # Spacing from smooth_header
coarse_space_directions = np.array(coarse_header['space directions']) # Space directions from coarse_header
# Calculate new space directions for smooth header
new_smooth_space_directions = np.copy(coarse_space_directions)
for i in range(len(smooth_spacing)):
norm_coarse = np.linalg.norm(coarse_space_directions[i])
new_smooth_space_directions[i] = smooth_spacing[i] * (coarse_space_directions[i] / norm_coarse)
new_smooth_header['space directions'] = new_smooth_space_directions
# # Assign the new header to the smooth image
# smooth_image_with_coarse_header = (smooth_image, new_smooth_header)
ordered_new_smooth_header = OrderedDict(new_smooth_header.items())
nrrd.write(path_to_save_image, smooth_image, header=ordered_new_smooth_header)
Now the new header is
OrderedDict([('type', 'uint8'),
('dimension', 3),
('space', 'left-posterior-superior'),
('sizes', array([1314, 884, 900])),
('space directions',
array([[ 0.13187834, -0.04589298, -0.05478995],
[ 0.05076999, 0.1410894 , 0.00402352],
[ 0.0503042 , -0.02208201, 0.13957748]])),
('kinds', ['domain', 'domain', 'domain']),
('encoding', 'raw'),
('space origin',
array([-148.47837839, -45.81777566, -68.16309759]))])
So in theory they should be in the same space, as far as I understand. They’re not. Size seems fine, as well as orientation, it looks that it just need a translation, but I don’t know which one.