Hi, I guess this is indeed a beginner question, but here I go.
I have a series of DICOM files of a Dixon sequence of the thigh. My first very simple task is to create a mask of the whole thigh against the background.
To do so at first I read a sitk.Image from DICOM and I use OtsuTreshold to get the outline of the thigh, then I use BinaryReconstructionByErosion in order to fill the outline and have the full mask.
This works fine when I do it in 2D (i.e. opening one DICOM at a time and processing it).
However, if I try to do it on a 3D image obtained by reading the entire DICOM series, the process fails, and instead of having a filled mask as output of the reconstruction I have a mask that is identical to the Otsu mask calculated in the first step (cfr plot below).
Here 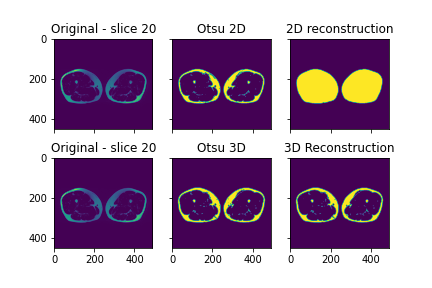
Here the code that I used:
from os import chdir
import SimpleITK as sitk
from glob import glob
import matplotlib.pyplot as plt
import numpy as np
from skimage.morphology import reconstruction
def copy_sitk_space_info(templateImage, newImage):
newImage.SetOrigin(templateImage.GetOrigin())
newImage.SetDirection(templateImage.GetDirection())
newImage.SetSpacing(templateImage.GetSpacing())
return(newImage)
def read_dicom_series(input_dcm_dir):
reader = sitk.ImageSeriesReader()
dicom_names = reader.GetGDCMSeriesFileNames(input_dcm_dir)
reader.SetFileNames(dicom_names)
inputImage = reader.Execute()
return(inputImage)
#input file
input_dcm_dir = "/my/dcm/dir"
chdir(input_dcm_dir)
f = glob("*")[20]
# 2D processing
im = sitk.ReadImage(f)
maskImage = sitk.OtsuThreshold( im, 0, 1, 200 )
plt.imshow(sitk.GetArrayFromImage(maskImage).squeeze())
mask_image_array = sitk.GetArrayFromImage(maskImage)
#initalise seed image
seed_image = np.zeros_like(mask_image_array)
seed_image[:,1:-1, 1:-1] = mask_image_array.max()
seedImage = sitk.GetImageFromArray(seed_image)
seedImage = copy_sitk_space_info(maskImage, seedImage)
filledImage = sitk.BinaryReconstructionByErosion(seedImage, maskImage)
#3D processing
inputImage = read_dicom_series(input_dcm_dir)
inputImage = sitk.Cast(inputImage, sitk.sitkFloat32)
maskImage_3D = sitk.OtsuThreshold( inputImage, 0, 1, 200 )
mask_image_array = sitk.GetArrayFromImage(maskImage_3D)
#initalise seed image
seed_image = np.zeros_like(mask_image_array)
seed_image[1:-1,1:-1, 1:-1] = mask_image_array.max()
seedImage = sitk.GetImageFromArray(seed_image)
seedImage = copy_sitk_space_info(maskImage_3D, seedImage)
filledImage3D = sitk.BinaryReconstructionByErosion(seedImage, maskImage_3D)
fig, axs = plt.subplots(2, 3)
axs[0, 0].imshow((sitk.GetArrayFromImage(im).squeeze()))
axs[0, 0].set_title('Original - slice 20')
axs[0, 1].imshow((sitk.GetArrayFromImage(maskImage).squeeze()))
axs[0, 1].set_title('Otsu 2D')
axs[0, 2].imshow((sitk.GetArrayFromImage(filledImage).squeeze()))
axs[0, 2].set_title('2D reconstruction')
axs[1, 0].imshow((sitk.GetArrayFromImage(im).squeeze()))
axs[1, 0].set_title('Original - slice 20')
axs[1, 1].imshow((sitk.GetArrayFromImage(maskImage_3D).squeeze()[20,:,:]))
axs[1, 1].set_title('Otsu 3D')
axs[1, 2].imshow((sitk.GetArrayFromImage(filledImage3D).squeeze()[20,:,:]))
axs[1, 2].set_title('3D Reconstruction')
for ax in axs.flat:
ax.label_outer()
plt.savefig("2D_3D_erosion.png")
I guess that my main question is: am I doing something wrong or does indeed the BinaryReconstructionByErosion only work on 2D images ? And if this is the case, what it the best and more adapted method to apply an operation to several slices of a 3D image and get back the output as another 3D image ?
Here you’ll find an archive with the original DICOM files that I used.
Thank in advance for any tips or suggestion