Hello @zivy,
Based on your suggestion, I am trying to convert the contour image to a distance map. However I do not know the code to do so. Please help.
I did not provide details because I have already faced the issues that you mentioned, earlier in this chain of question answers. So I assume these issues are already taken care of. However, below is the code and the relevant screenshots. Please let me know if there is anything erroneous.
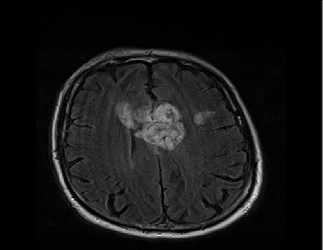
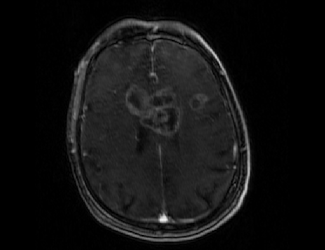
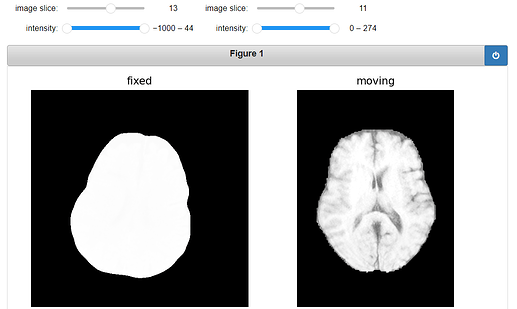
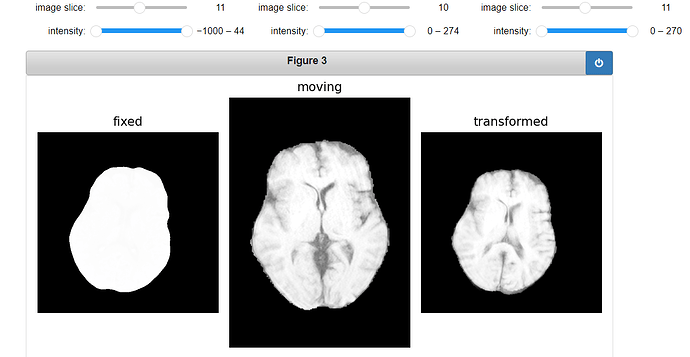
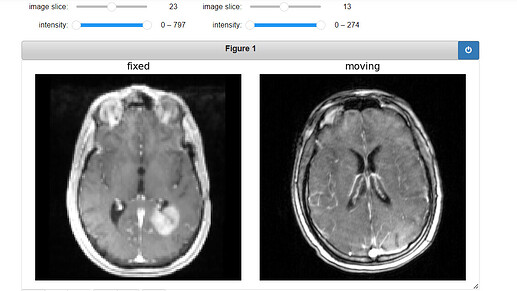
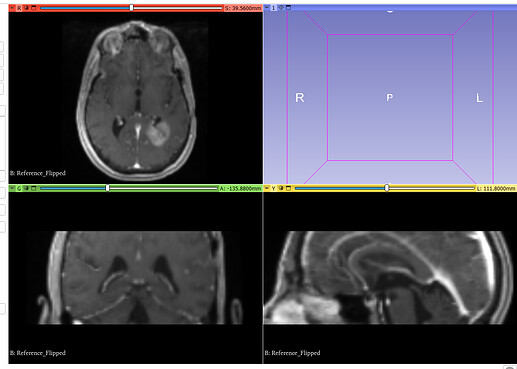
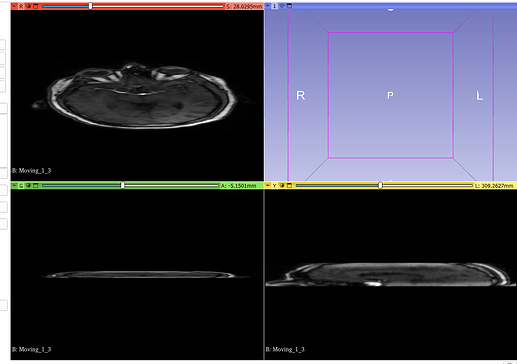
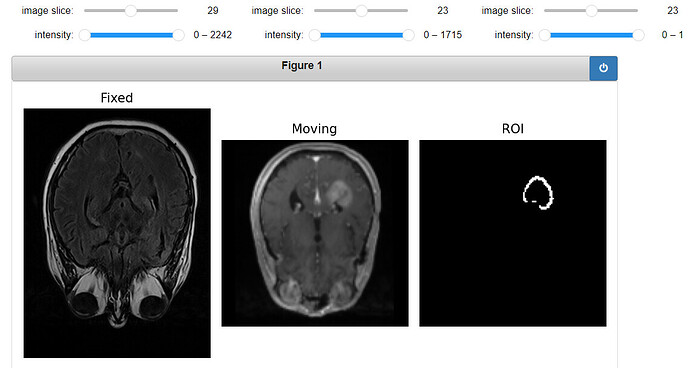
Original Fixed, Moving and ROI image:
Registration Code:
initial_transform = sitk.CenteredTransformInitializer(
fixed_image,
moving_image,
sitk.Euler3DTransform(),
#sitk.ComposeScaleSkewVersor3DTransform(),
sitk.CenteredTransformInitializerFilter.GEOMETRY,
)
registration_method = sitk.ImageRegistrationMethod()
# Similarity metric settings.
registration_method.SetMetricAsMattesMutualInformation(numberOfHistogramBins=50)
registration_method.SetMetricSamplingStrategy(registration_method.RANDOM)
registration_method.SetMetricSamplingPercentage(0.1)
registration_method.SetInterpolator(sitk.sitkLinear)
#registration_method.SetInterpolator(sitk.sitkNearestNeighbor)
# Optimizer settings.
registration_method.SetOptimizerAsGradientDescent(
learningRate=1.0,
numberOfIterations=1000,
convergenceMinimumValue=1e-6,
convergenceWindowSize=10,
)
registration_method.SetOptimizerScalesFromPhysicalShift()
# Setup for the multi-resolution framework.
# registration_method.SetShrinkFactorsPerLevel(shrinkFactors = [4,2,1])
# registration_method.SetSmoothingSigmasPerLevel(smoothingSigmas=[2,1,0])
# registration_method.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
# Don't optimize in-place, we would possibly like to run this cell multiple times.
registration_method.SetInitialTransform(initial_transform, inPlace=False)
#Connect all of the observers so that we can perform plotting during registration.
registration_method.AddCommand(sitk.sitkStartEvent, rgui.start_plot)
registration_method.AddCommand(sitk.sitkEndEvent, rgui.end_plot)
registration_method.AddCommand(
sitk.sitkMultiResolutionIterationEvent, rgui.update_multires_iterations
)
registration_method.AddCommand(
sitk.sitkIterationEvent, lambda: rgui.plot_values(registration_method)
)
final_transform = registration_method.Execute(fixed_image, moving_image)
# Always check the reason optimization terminated.
print("Final metric value: {0}".format(registration_method.GetMetricValue()))
print(
"Optimizer's stopping condition, {0}".format(
registration_method.GetOptimizerStopConditionDescription()
)
)
moving_resampled = sitk.Resample(
moving_image,
fixed_image,
final_transform,
sitk.sitkLinear,
0.0,
moving_image.GetPixelID(),
)
roi_resampled = sitk.Resample(
ROI_image,
fixed_image,
final_transform,
#sitk.sitkLinear,
#sitk.sitkNearestNeighbor,
sitk.sitkLabelGaussian,
0.0,
ROI_image.GetPixelID(),
)
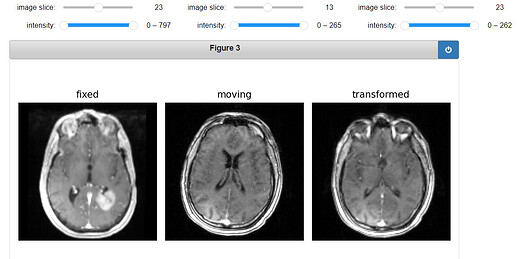
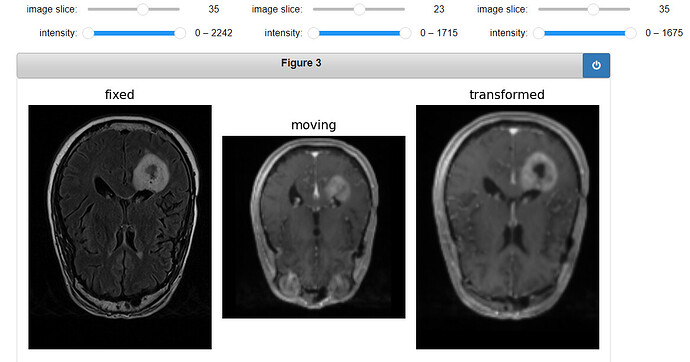
gui.MultiImageDisplay(
image_list=[fixed_image, moving_image, moving_resampled],
title_list=["fixed", "moving", "transformed"],
figure_size=(8, 4),
window_level_list=[fixed_window_level, moving_window_level, moving_window_level],
);
Output:
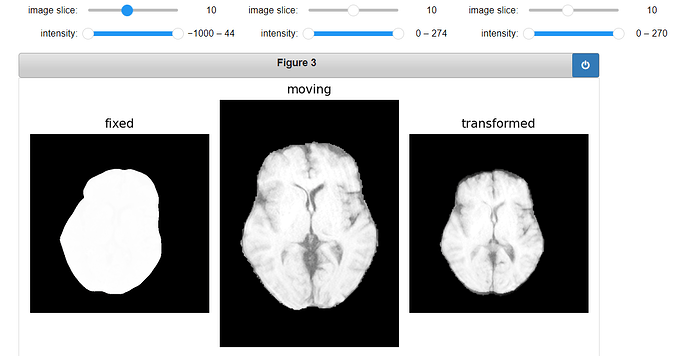
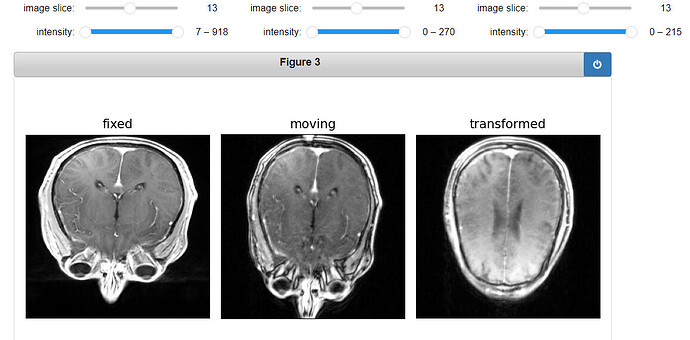
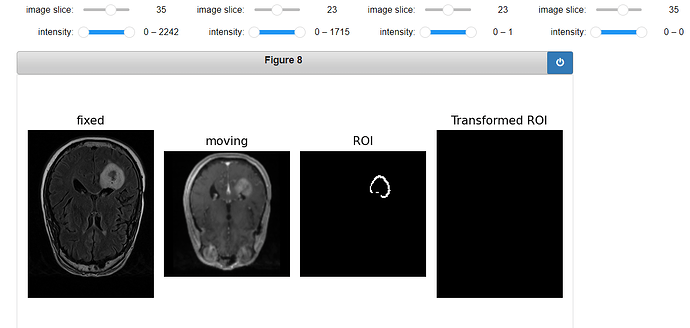
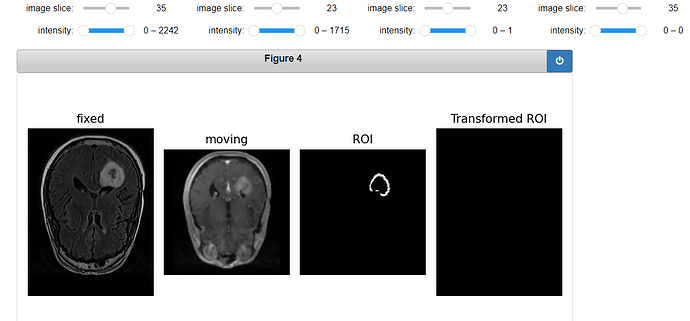
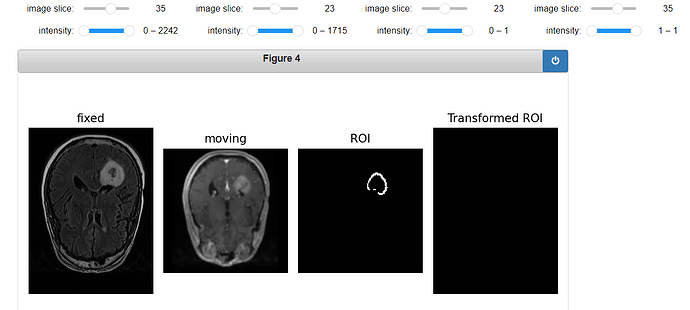
Code for displaying the transformed ROI:
gui.MultiImageDisplay(
image_list=[fixed_image, moving_image, ROI_image, roi_resampled],
title_list=["fixed", "moving", "ROI", "Transformed ROI"],
figure_size=(8, 4),
window_level_list=[fixed_window_level, moving_window_level, ROI_window_level, ROI_window_level],
);
Output