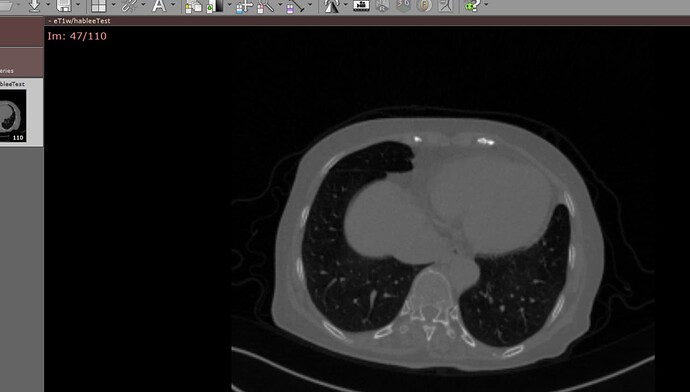
hi, I tried to read dicom files which are all stored in a directory using the following code
void readDicomSeries(const std::string& dicom_dir, itk::Image<float, 3>::Pointer& out_image)
{
using ImageIOType = itk::GDCMImageIO;
ImageIOType::Pointer DicomIO = ImageIOType::New();
using ImageReaderType3D = itk::ImageSeriesReader<itk::Image<float, 3>>;
ImageReaderType3D::Pointer ImageReader3D = ImageReaderType3D::New();
ImageReader3D->SetImageIO(DicomIO);
using NamesGeneratorType = itk::GDCMSeriesFileNames;
NamesGeneratorType::Pointer NameGenerator = NamesGeneratorType::New();
NameGenerator->SetDirectory(dicom_dir);
using SeriesIdContainer = std::vector<std::string>;
const SeriesIdContainer& SeriesUID = NameGenerator->GetSeriesUIDs();
SeriesIdContainer::const_iterator SeriesItr_begin = SeriesUID.begin();
SeriesIdContainer::const_iterator SeriesItr_end = SeriesUID.end();
while (SeriesItr_begin != SeriesItr_end)
{
std::cout << SeriesItr_begin->c_str() << std::endl;
SeriesItr_begin++;
}
std::string SeriesIdentifier;
SeriesIdentifier = SeriesUID.begin()->c_str();
using FileNamesContainer = std::vector<std::string>;
FileNamesContainer filenames;
filenames = NameGenerator->GetFileNames(SeriesIdentifier);
ImageReader3D->SetFileNames(filenames);
try
{
ImageReader3D->Update();
}
catch (itk::ExceptionObject& err)
{
std::cerr << "error, exception object caught!" << std::endl;
}
out_image = ImageReader3D->GetOutput();
}
the files looks right, and I can open them with radiAnt software, but I got the exception that saying “GDCMSeriesFileNames (000001DDACC3E6C0): No Series were found”.
all files looks like this:
how can I solve the problem? Thank you!