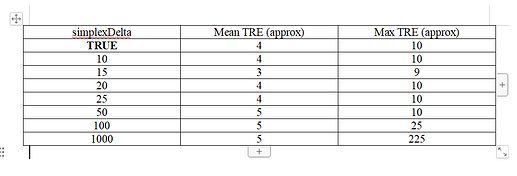
A few simplexDelta values, and the corresponding Mean TRE and Max TRE values obtained, are listed below.
However, each time, the code stopped with the following exception
Unexpected exception formatting exception. Falling back to standard exception
Traceback (most recent call last):
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\interactiveshell.py", line 3548, in run_code
exec(code_obj, self.user_global_ns, self.user_ns)
File "C:\Users\debap\AppData\Local\Temp\ipykernel_19716\1123557497.py", line 1, in <module>
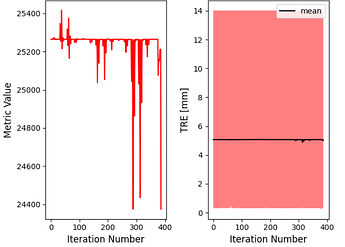
get_ipython().run_cell_magic('timeit', '-r1 -n1', '\nglobal tx\n\n# Select the fixed and moving images, valid entries are in [0,9].\n# fixed_image_index = 0\n# moving_image_index = 7\n\n# fixed_image_index = 1\n# moving_image_index = 8\n\n# fixed_image_index = 2\n# moving_image_index = 9\n\n# fixed_image_index = 0\n# moving_image_index = 9\n\n# fixed_image_index = 0\n# moving_image_index = 8\n\n# fixed_image_index = 3\n# moving_image_index = 6\n\n# fixed_image_index = 1\n# moving_image_index = 6\n\n# fixed_image_index = 1\n# moving_image_index = 7\n\n# fixed_image_index = 1\n# moving_image_index = 5\n\nfixed_image_index = 5\nmoving_image_index = 9\n\n\n\ntx = bspline_intra_modal_registration(\n fixed_image=images[fixed_image_index],\n moving_image=images[moving_image_index],\n fixed_image_mask=(masks[fixed_image_index] == lung_label),\n fixed_points=points[fixed_image_index],\n moving_points=points[moving_image_index],\n)\n(\n initial_errors_mean,\n initial_errors_std,\n _,\n initial_errors_max,\n initial_errors,\n) = ru.registration_errors(\n sitk.Euler3DTransform(), points[fixed_image_index], points[moving_image_index]\n)\n(\n final_errors_mean,\n final_errors_std,\n _,\n final_errors_max,\n final_errors,\n) = ru.registration_errors(tx, points[fixed_image_index], points[moving_image_index])\n\nplt.hist(initial_errors, bins=500, alpha=0.5, label="before registration", color="blue")\nplt.hist(final_errors, bins=500, alpha=0.5, label="after registration", color="green")\nplt.legend()\nplt.title("TRE histogram")\nprint(\n f"Initial alignment errors in millimeters, mean(std): {initial_errors_mean:.2f}({initial_errors_std:.2f}), max: {initial_errors_max:.2f}"\n)\nprint(\n f"Final alignment errors in millimeters, mean(std): {final_errors_mean:.2f}({final_errors_std:.2f}), max: {final_errors_max:.2f}"\n)\n\n\n# print(f"{initial_errors_med:.2f}")\n# print(f"{final_errors_med:.2f}")\n')
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\interactiveshell.py", line 2515, in run_cell_magic
result = fn(*args, **kwargs)
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\magics\execution.py", line 1189, in timeit
all_runs = timer.repeat(repeat, number)
File "C:\Users\debap\AppData\Local\Programs\Python\Python310\lib\timeit.py", line 206, in repeat
t = self.timeit(number)
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\magics\execution.py", line 173, in timeit
timing = self.inner(it, self.timer)
File "<magic-timeit>", line 36, in inner
File "C:\Users\debap\AppData\Local\Temp\ipykernel_19716\2841005318.py", line 78, in bspline_intra_modal_registration
return registration_method.Execute(fixed_image, moving_image)
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\SimpleITK\SimpleITK.py", line 10863, in Execute
val = _SimpleITK.ImageRegistrationMethod_Execute(self, fixed, moving)
RuntimeError: Exception thrown in SimpleITK ImageRegistrationMethod_Execute: C:\Users\debap\Desktop\ITK\Modules\Numerics\Optimizersv4\src\itkAmoebaOptimizerv4.cxx:268:
ITK ERROR: AmoebaOptimizerv4(000002A650BF1D80): cost function and simplex delta dimensions mismatch
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\interactiveshell.py", line 2142, in showtraceback
stb = self.InteractiveTB.structured_traceback(
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\ultratb.py", line 1435, in structured_traceback
return FormattedTB.structured_traceback(
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\ultratb.py", line 1326, in structured_traceback
return VerboseTB.structured_traceback(
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\ultratb.py", line 1173, in structured_traceback
formatted_exception = self.format_exception_as_a_whole(etype, evalue, etb, number_of_lines_of_context,
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\ultratb.py", line 1063, in format_exception_as_a_whole
self.get_records(etb, number_of_lines_of_context, tb_offset) if etb else []
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\ultratb.py", line 1155, in get_records
FrameInfo(
File "C:\Users\debap\Desktop\Python_Vir_Env\Python\lib\site-packages\IPython\core\ultratb.py", line 780, in __init__
ix = inspect.getsourcelines(frame)
File "C:\Users\debap\AppData\Local\Programs\Python\Python310\lib\inspect.py", line 1129, in getsourcelines
lines, lnum = findsource(object)
File "C:\Users\debap\AppData\Local\Programs\Python\Python310\lib\inspect.py", line 958, in findsource
raise OSError('could not get source code')
OSError: could not get source code
Can you please share an example code which uses the Amoeba optimizer?