I’ve tried to break this down into two main issues and have provided code and de-identified data as well. For reference, the machine is a GE Lightspeed VCT and the study is a triple-phase CT AP. Nothing turned up on Google search of dicom issues with that scanner… but that probably doesn’t mean much.
https://www.dropbox.com/sh/tcnq9bdso7s046a/AACWmSvv5jaCLKJ33_WsZFFZa?dl=0 (CLEAN DICOM DATA)
First Problem: Multiple Volumes Per Series Using GetGDCMSeriesFileNames.
And when using GetGDCMSeriesFileNames with useSeriesDetails it generates an empty list  . With plain old GetGDCMSeriesFileNames, as in code below, the STANDARD RECON series includes multiple volumes, specifically multiple contrast phase acquisitions in one series.
. With plain old GetGDCMSeriesFileNames, as in code below, the STANDARD RECON series includes multiple volumes, specifically multiple contrast phase acquisitions in one series.
data_directory = "1.2.840.113619.2.334.3.2831163449.135.1463999344.729"
out_dir = "./useSeriesDetailsTest"
series_IDs = sitk.ImageSeriesReader.GetGDCMSeriesIDs(data_directory)
if not series_IDs:
print("ERROR: given directory \""+data_directory+"\" does not contain a DICOM series.")
sys.exit(1)
for i,series_ID in enumerate(series_IDs):
series_file_names = sitk.ImageSeriesReader.GetGDCMSeriesFileNames(data_directory,
series_ID,useSeriesDetails=False) #useSeriesDetails ?
series_reader = sitk.ImageSeriesReader()
series_reader.SetFileNames(series_file_names)
try:
img = series_reader.Execute()
except RuntimeError:
print ("--> Fundamental error in image layer, skipping...")
continue
nreader = sitk.ReadImage(series_file_names[0])
print (str(nreader.GetMetaData("0008|103e"))+" is being processed")
series_desc = nreader.GetMetaData("0008|103e")
sitk.WriteImage(img,os.path.join(out_dir,str(i)+"_"+str(series_desc)+".nii.gz"))
Printing out dicom tags for slice position, series description, acquisition time and series uid (in that order) we see why this is. A single series UID is associated with data from different time points–clinically these are two different contrast phases.
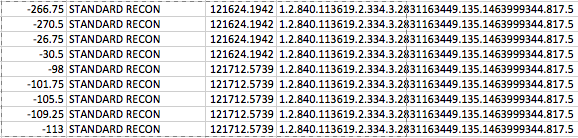
Problem Two: Out of Order Slices When Applying Work-around That Feeds Only DCMs From Single Volume
To combat problem one I wrote code for a work around that is included earlier in this thread. Essentially, it makes a list of all dcms as shown above, then groups file paths for dcms with the same acquisition time AND series uid. This works to isolate dcms associated with a single volume, but when these lists are then fed back into SimpleITK to generate a volume some are out of order (…and some work). And those out of order are not randomly out of order…but not consistently out of order in the same way either. See an example here:
And when I drag only those dcms associated with single volume into Slicer or Osirix they magically appear in the right order!! So there may be something unique to SimpleITK causing this…
In the meantime, any suggestions on an alternate dicom to nifti conversion tool would be greatly appreciated as I already have this “sorting dcms by series” solution above.
Apologies for the insanely long post. SimpleITK has been a cornerstone for tons of my work and greatly appreciate all that you guys do. I hope this might help your efforts in some way.
-Brett
 . With plain old GetGDCMSeriesFileNames, as in code below, the STANDARD RECON series includes multiple volumes, specifically multiple contrast phase acquisitions in one series.
. With plain old GetGDCMSeriesFileNames, as in code below, the STANDARD RECON series includes multiple volumes, specifically multiple contrast phase acquisitions in one series.