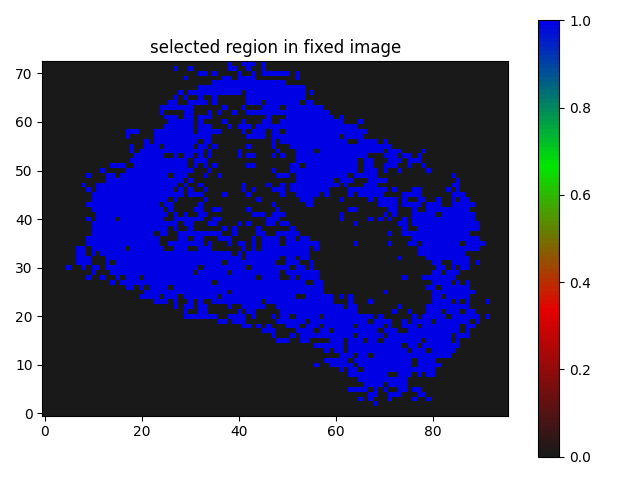
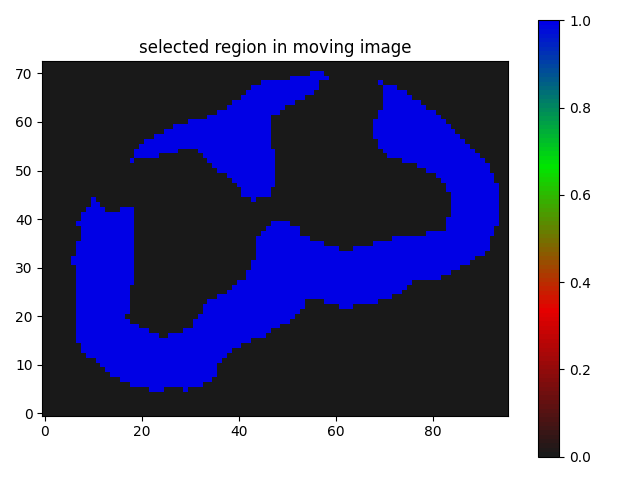
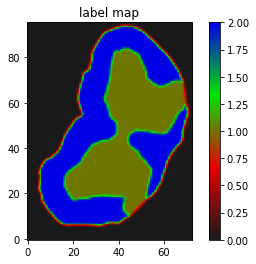
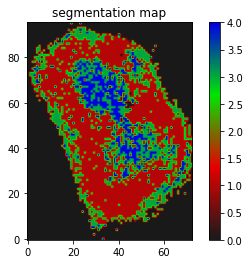
I have a mask that has 2 labels. I want to register it on a segmentation map that has 3 labels(both cases background 0).
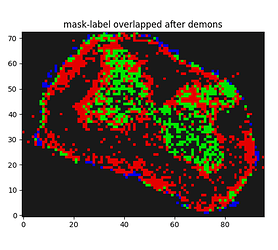
I have run the following method but getting bad result outputs:
mask_itk = sitk.GetImageFromArray(mask)
label_itk = sitk.GetImageFromArray(labelMap)
fixed_image = sitk.Cast(label_itk, sitk.sitkFloat32)
moving_image = sitk.Cast(mask_itk, sitk.sitkFloat32)
initialTx = sitk.CenteredTransformInitializer(fixed_image, moving_image, sitk.AffineTransform(fixed_image.GetDimension()))
registrationMethod = sitk.ImageRegistrationMethod()
# registrationMethod.SetShrinkFactorsPerLevel([1, 1, 1])
# registrationMethod.SetSmoothingSigmasPerLevel([1, 1, 1])
# registrationMethod.SetMetricAsJointHistogramMutualInformation(20)
registrationMethod.MetricUseFixedImageGradientFilterOff()
registrationMethod.SetOptimizerAsGradientDescent(
learningRate=10.0,
numberOfIterations=1000,
estimateLearningRate=registrationMethod.EachIteration,
)
registrationMethod.SetOptimizerScalesFromPhysicalShift()
registrationMethod.SetInitialTransform(initialTx)
registrationMethod.SetInterpolator(sitk.sitkLinear)
registrationMethod.AddCommand(sitk.sitkIterationEvent, lambda: command_iteration(registrationMethod))
registrationMethod.AddCommand(sitk.sitkMultiResolutionIterationEvent,
lambda: command_multiresolution_iteration(registrationMethod))
outTx1 = registrationMethod.Execute(fixed_image, moving_image)
print("-------")
print(outTx1)
print(f"Optimizer stop condition: {registrationMethod.GetOptimizerStopConditionDescription()}")
print(f" Iteration: {registrationMethod.GetOptimizerIteration()}")
print(f" Metric value: {registrationMethod.GetMetricValue()}")
displacementField = sitk.Image(fixed_image.GetSize(), sitk.sitkVectorFloat64)
displacementField.CopyInformation(fixed_image)
displacementTx = sitk.DisplacementFieldTransform(displacementField)
# del displacementField
displacementTx.SetSmoothingGaussianOnUpdate(
varianceForUpdateField=0.0, varianceForTotalField=1.5
)
registrationMethod.SetMovingInitialTransform(outTx1)
registrationMethod.SetInitialTransform(displacementTx, inPlace=True)
registrationMethod.SetMetricAsANTSNeighborhoodCorrelation(4)
registrationMethod.MetricUseFixedImageGradientFilterOff()
# registrationMethod.SetShrinkFactorsPerLevel([3, 2, 1])
# registrationMethod.SetSmoothingSigmasPerLevel([2, 1, 1])
registrationMethod.SetOptimizerScalesFromPhysicalShift()
registrationMethod.SetOptimizerAsGradientDescent(
learningRate=1,
numberOfIterations=300,
estimateLearningRate=registrationMethod.EachIteration,
)
registrationMethod.Execute(fixed_image, moving_image)
print("-------")
print(displacementTx)
print(f"Optimizer stop condition: {registrationMethod.GetOptimizerStopConditionDescription()}")
print(f" Iteration: {registrationMethod.GetOptimizerIteration()}")
print(f" Metric value: {registrationMethod.GetMetricValue()}")
compositeTx = sitk.CompositeTransform([outTx1, displacementTx])
resampler = sitk.ResampleImageFilter()
resampler.SetReferenceImage(fixed_image)
resampler.SetInterpolator(sitk.sitkLinear)
resampler.SetDefaultPixelValue(0)
resampler.SetTransform(compositeTx)
out = resampler.Execute(moving_image)
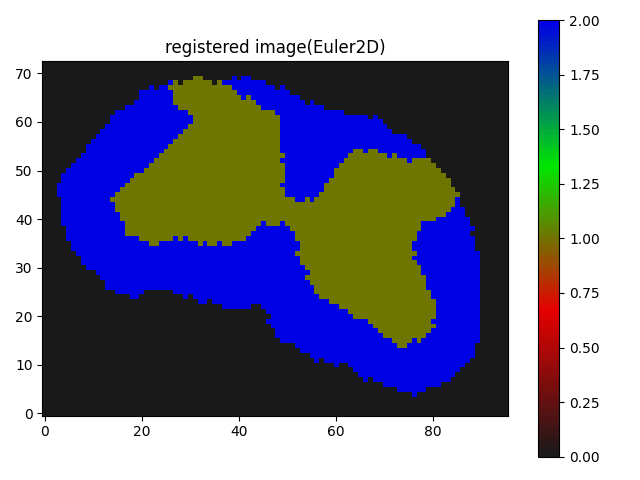
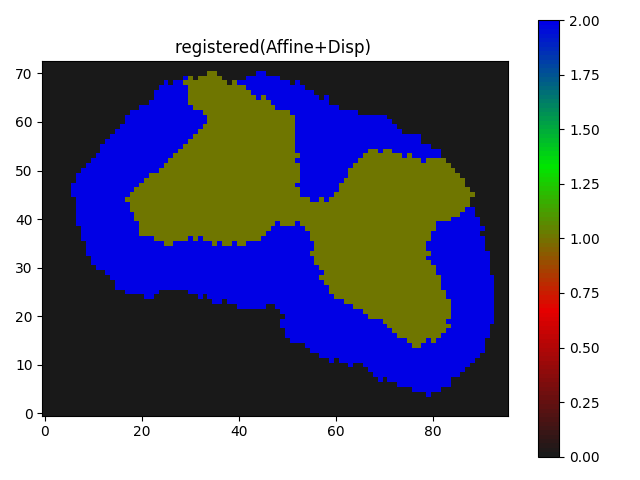
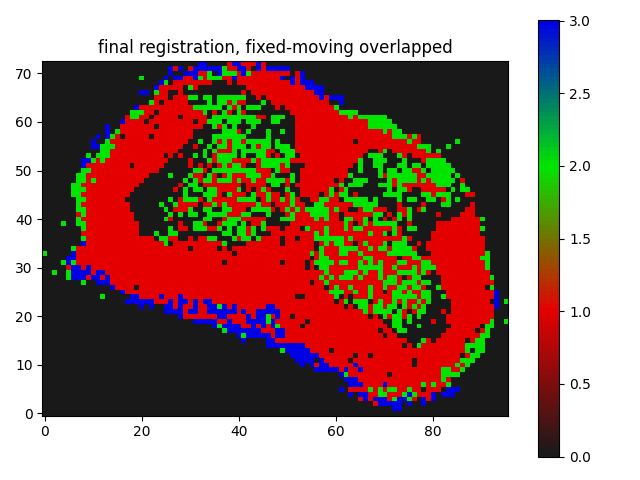
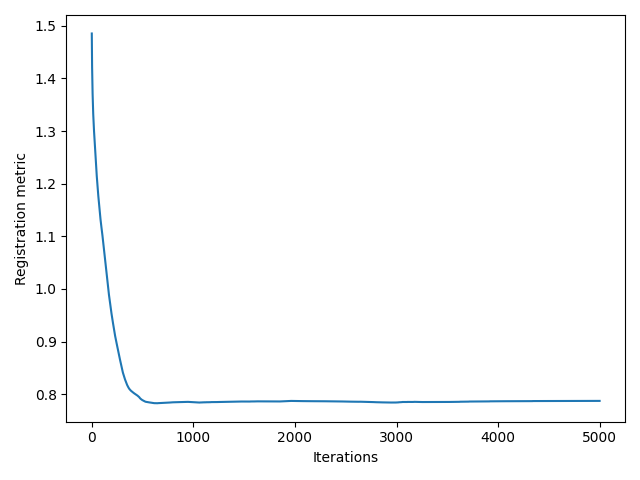
The result I am getting is as follows:
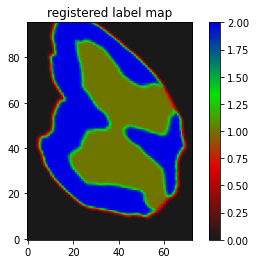
Though I believe it is somewhat working but I guess not the kind of registration results I need, where the butterfly regions and the red parts of the segmentation should match/overlap.
Any suggestions, or ideas to improve would be very appreciated.
Thanks.
seg_label_map.h5 (84.1 KB)
The sample data both label and segmentation map can be found above. Here is the code.
!pip install h5py
import os
pathToTheFile= os.path.join(downloadDir, 'seg_label_map.h5')
with h5py.File(reconPath, 'r') as pfile:
segmentation = np.array(pfile['segmentation'])
label = np.array(pfile['label'])
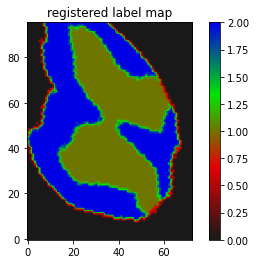
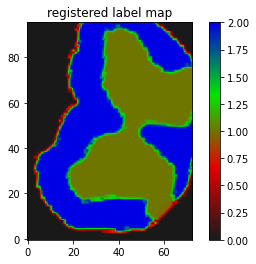
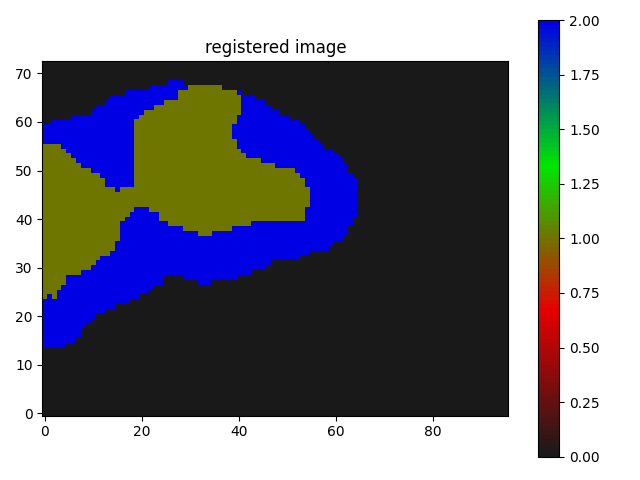
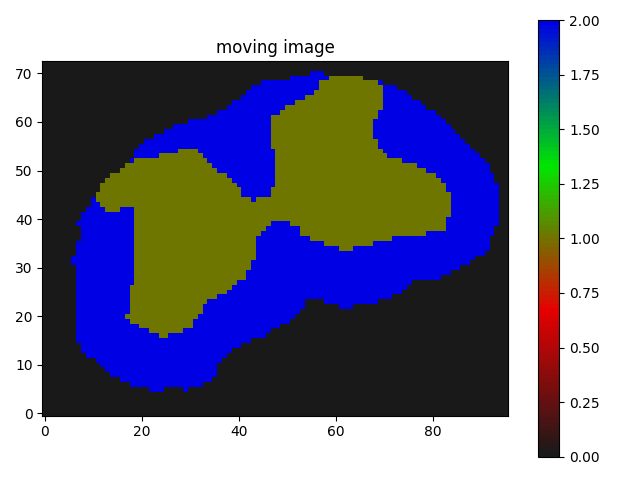 just translated I suppose.
just translated I suppose.