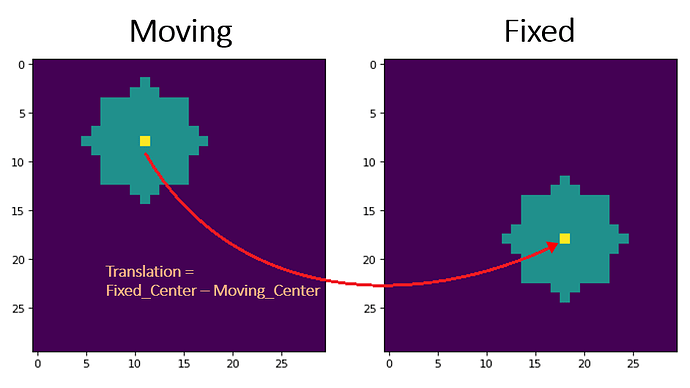
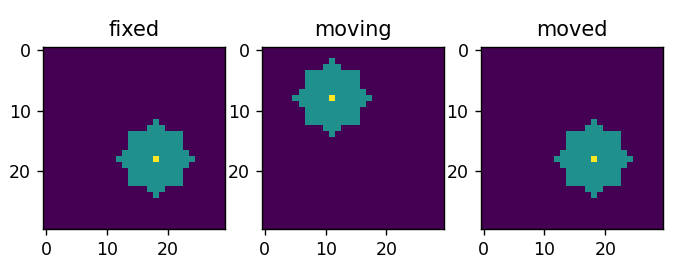
Hi!
I hope I am putting this question in the proper category.
I followed the Jupyter notebook concerning the 3D registration and I tried to apply it to my problem.
Here is my problem: I have 2 axial series, 1 fixed and 1 moving. The idea is to click on a voxel of the moving series and to display the equivalent position in the fixed series.
To do so, I decided to take the moving series and register it onto the fixed series. That way I obtain a transformation that is able to compute the voxel position of the moving series to the fixed series.
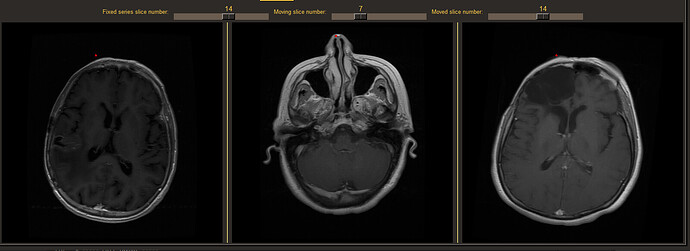
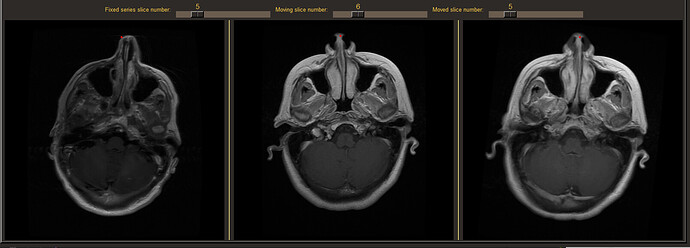
Here is an example:
From left to right, you can see, the fixed series (left), the moving series (middle) and the moved series (right).
The problem I have is when i click on the nose of the moving series (middle), the computation of the location of the pixel on the fixed series is erroneous.
- Moving point [241, 31, 6]
- Moved point [211, 73, 13]
I would have expected the moved point to be [280, 60, 4]
I suppose I made a mistake but I cannot see it. Therefore, I need you help determining how I could fix it.
Here is the full code:
import SimpleITK as sitk
def registration_3d(fixed_image, moving_image):
# Initial Alignment
# Use the CenteredTransformInitializer to align the centers of the two volumes and set the center of rotation to the center of the fixed image
initial_transform = sitk.CenteredTransformInitializer(
fixed_image,
moving_image,
sitk.Euler3DTransform(),
sitk.CenteredTransformInitializerFilter.GEOMETRY,
)
# Registration
registration_method = sitk.ImageRegistrationMethod()
# Similarity metric settings.
registration_method.SetMetricAsMattesMutualInformation(numberOfHistogramBins=50)
registration_method.SetMetricSamplingStrategy(registration_method.RANDOM)
registration_method.SetMetricSamplingPercentage(0.01)
registration_method.SetInterpolator(sitk.sitkLinear)
# Optimizer settings.
registration_method.SetOptimizerAsGradientDescent(
learningRate=0.5,
numberOfIterations=100,
convergenceMinimumValue=1e-10,
convergenceWindowSize=10,
)
registration_method.SetOptimizerScalesFromPhysicalShift()
# Setup for the multi-resolution framework.
registration_method.SetShrinkFactorsPerLevel(shrinkFactors=[4, 2, 1])
registration_method.SetSmoothingSigmasPerLevel(smoothingSigmas=[2, 1, 0])
registration_method.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
# Don't optimize in-place, we would possibly like to run this cell multiple times.
# registration_method.SetInitialTransform(initial_transform, inPlace=False)
registration_method.SetInitialTransform(initial_transform, inPlace=False)
final_transform = registration_method.Execute(
sitk.Cast(fixed_image, sitk.sitkFloat32), sitk.Cast(moving_image, sitk.sitkFloat32)
)
# Post registration analysis
print(f"Final metric value: {registration_method.GetMetricValue()}")
print(
f"Optimizer's stopping condition, {registration_method.GetOptimizerStopConditionDescription()}"
)
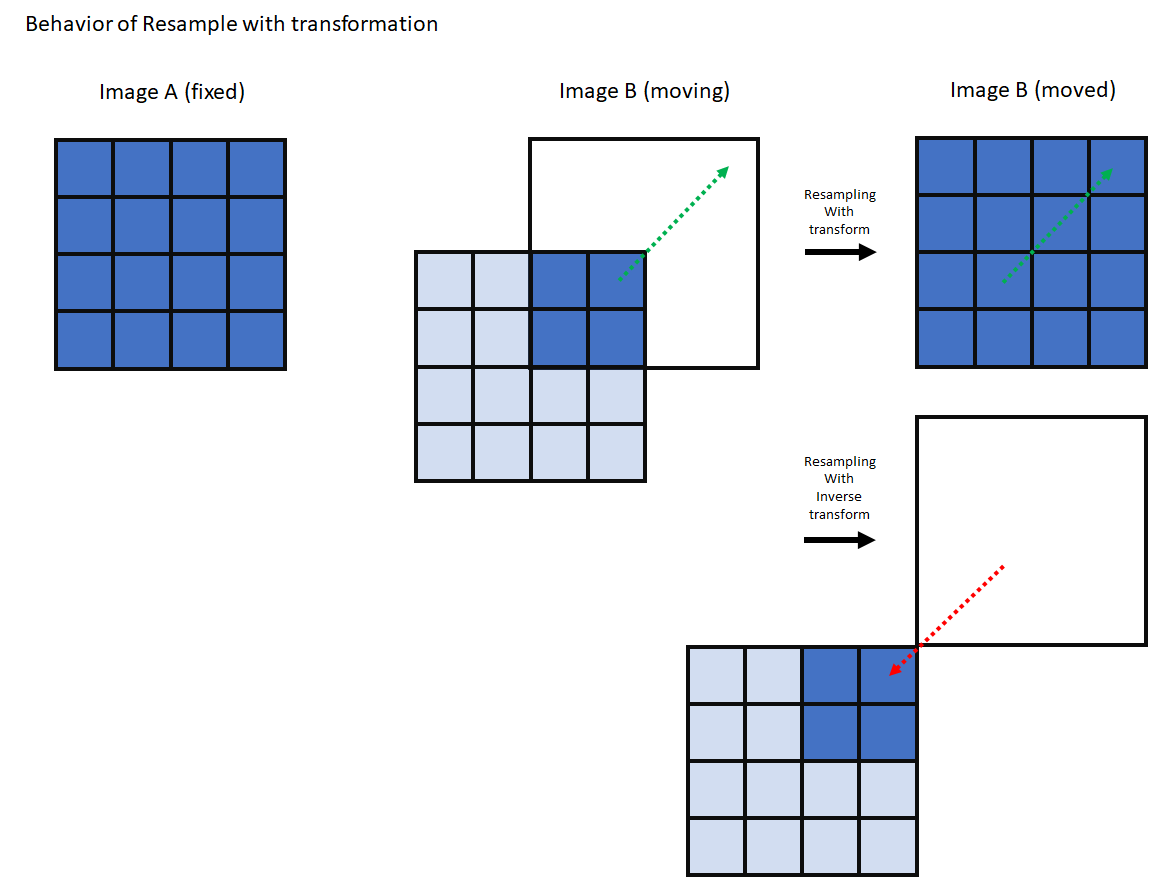
moving_resampled = sitk.Resample(
moving_image,
fixed_image,
final_transform,
sitk.sitkLinear,
0.0,
moving_image.GetPixelID(),
)
return moving_resampled, [initial_transform, final_transform]
reader = sitk.ImageSeriesReader()
dicom_files = reader.GetGDCMSeriesFileNames("C:/test_dirs/DCM/ACQ1_24img")
reader.SetFileNames(dicom_files)
fixed_image = reader.Execute()
fixed_image = sitk.Cast(fixed_image, sitk.sitkFloat32)
reader = sitk.ImageSeriesReader()
dicom_files = reader.GetGDCMSeriesFileNames("C:/test_dirs/DCM/ACQ1_24img_2")
reader.SetFileNames(dicom_files)
moving_image = reader.Execute()
moving_image = sitk.Cast(moving_image, sitk.sitkFloat32)
moved, [init_transform, final_transform] = registration_3d(fixed_image, moving_image)
sitk.Show(fixed_image, "Fixed")
sitk.Show(moving_image, "Moving")
sitk.Show(moved, "Moved")
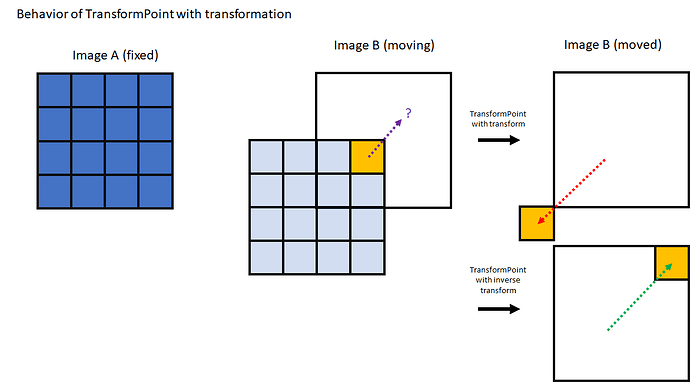
point_init_index = [241, 31, 6]
p_init = moving_image.TransformContinuousIndexToPhysicalPoint([point_init_index[0], point_init_index[1], point_init_index[2]])
p_final = final_transform.TransformPoint(p_init)
p_final_index = moved.TransformPhysicalPointToIndex(p_final)
print(f'Moving point {point_init_index}')
print(f'Moved point {p_final_index}')
Thanks in advance!
PS: I do not know how to upload series if you needed to test with my data. Do not hesitate to tell me how to do it if needed.