I want to registration two images by SimpleITK. The image1 is axially acquired, while the image2 is coronary acquired. I find the result is poor. I wonder if it is possible to adjust the image1 and image2 by the patient orientation before registration? How can I implement this function by SimpleITK?
Hello @zhang-qiang-github,
Based on the information you provided, it is hard to tell what is the issue, not enough information.
Generally speaking, the acquisition (axial or coronal) is implicitly accounted for, as both images are read as 3D volumes. A critical aspect of registration is initialization. Is the initial alignment close? Is there a significant rotational or scale difference between the two images after initialization?
Also, possibly provide some information with respect to the image geometry: sizes, spacings and content: is this a sparse image with content only at a few locations or is there content throughout the volume?
For fixed image:
size: 500, 640, 243
spacing: 0.328 0.328 0.400
direction:
0.9997 0.022687 0
-0.022687 0.999743 0
0 0 1
for moving image:
size: 448 448 352
spacing: 0.404 0.404 0.4
direction:
5.4825e-5 0.0314 -0.9995
0.999998 -0.001745 3.834e-9
-0.00174 -0.9995 -0.03141
and the initial transform is:
initial_transform = sitk.CenteredTransformInitializer(fixed_image,
moving_image,
sitk.Euler3DTransform(),
sitk.CenteredTransformInitializerFilter.GEOMETRY)
itk::simple::Transform
Euler3DTransform (000002198E2E2140)
RTTI typeinfo: class itk::Euler3DTransform<double>
Reference Count: 1
Modified Time: 6885049
Debug: Off
Object Name:
Observers:
none
Matrix:
1 0 0
0 1 0
0 0 1
Offset: [-139.597, -0.620332, -3.92181]
Center: [0.98381, -27.831, 12.2481]
Translation: [-139.597, -0.620332, -3.92181]
Inverse:
1 0 0
0 1 0
0 0 1
Singular: 0
Euler's angles: AngleX=0 AngleY=0 AngleZ=0
m_ComputeZYX = 0
Finally, the registration result is:
for fixed image:
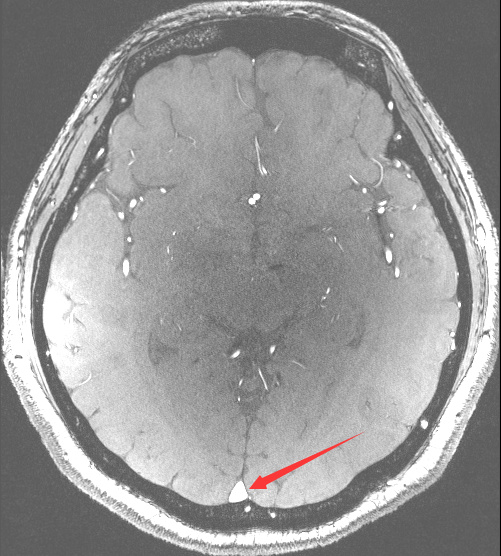
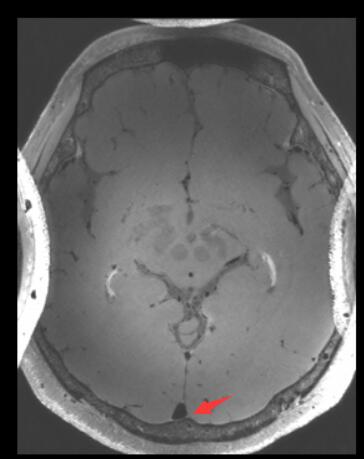
the image after registration
The red arrow point out that the registration result is upside down. It make me very confuzed.
Hello @zhang-qiang-github,
How does the resampled moving image look like after applying the initial transform? I suspect that it is 180 degrees rotated from what it should be. If the rotational difference between the two images is very large, registration will fail.
Please take a look at this Jupyter notebook, it illustrates a similar situation and how to resolve it.
The image is upside down after applying the initial transform. Is the initial transform calculated by the world coordinate system? Why the image is 180 degrees rotated from what it should be?
Hello @zhang-qiang-github,
As described in the referenced notebook, this can be due to human error when entering the patient position in the scanner. Real world data is not always clean and correct.
When you are using the CenteredTransformInitializer you are assuming that the rotation is not that significant. This assumption does not seem to hold in your case, which means you need to use a slightly more elaborate initialization strategy, as shown in the referenced notebook. Once you have a reasonable initialization, the registration will most succeed.
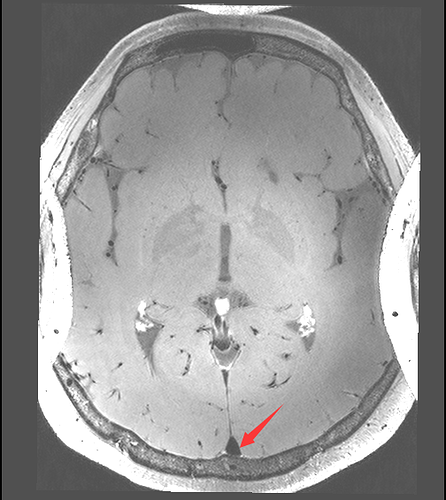
Dear zivy, I find one strange thing. As the above description, the registration result is upside down. However, when I use radiant to show the image, after show the coronal image as axial, they are almost registrated.
Please have a look at following picture:
Does it mean that the CenteredTransformInitializer should achieve the same result as RadiAnt? Is this a bug for SimpleITK?
Hello @zhang-qiang-github,
Not sure what it is you are asking. SimpleITK does not do display, so after obtaining a transformation with the CenteredTransformInitializer are you resampling the moving image onto the fixed image grid and saving to disk, and then displaying that with RadiAnt? If yes, and the display looks good then all is well, and the problem was with the display you previously used (SimpleITK doesn’t have built in display capabilities so it is always an external program or minimal code that is used).
If this initial registration looks reasonable, please run the full registration, do the resampling afterwards, and visualize the resulting image with RadiAnt to judge if your result is reasonable.
-
For the original images, I show it with RadiAnt. After showing the coronal image as axial, the two images looks good.
-
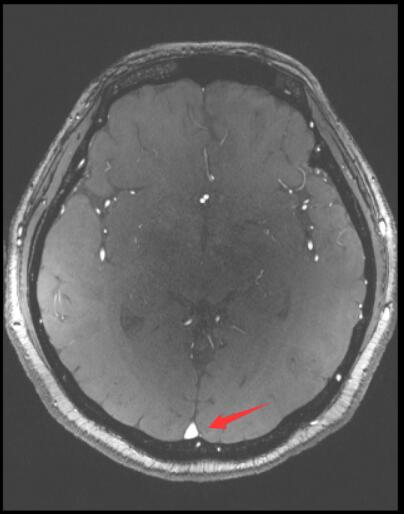
In SimpleITK, I resample the moving image onto the fixed image grid. Then, I get the numpy matrix from the resampled image and show it. It shows the upside down image.
Please follow the same path for both. Write the resampled moving image to disk and use RadiAnt to view it. If the problem is gone, then it was an issue with the numpy data display and not the data/registration itself. Otherwise there is an issue with the data/registration and we need to investigate that (a possible issue with SimpleITK or registration initialization).