I’m using Similarity2DTransform on binary image masks to pre-align the images before performing a B-spline registration step. In the majority of the images it works fine but there are a few examples where the Similarity2DTransform behaves weirdly.
Or better to say: the combination of image size, image spacing, optimizier parameters, interpolation and metric sometimes results in a complete mis-registration.
And it seems that everything is very sensitive to each of those parameters. In some cases there are only a very small range of parameters really working.
All the images are to some extend pre-aligned already and do overlap.
I could also create some test images to show the behaviour.
For example choosing some parameters results in a quite good registration
ConjugateGradientLineSearchOptimizerv4Template: Convergence checker passed at iteration 77.
But, If I change the parameters, for example setting the line search upper limit to 2.0, the rotation is gone:
ConjugateGradientLineSearchOptimizerv4Template: Convergence checker passed at iteration 14.
But it get’s even weirder if I set the metric sampling percentage to 100%:
ConjugateGradientLineSearchOptimizerv4Template: Convergence checker passed at iteration 19.
Now look at the iterations:
0 = 3737.46839: (1.0104391011536373, 0.004580075860924656, 0.44180988995370674, 2.5672543179081515)
1 = 3512.12638: (1.0256760248048011, 0.016888045821357972, -0.4908690733221168, 7.108722357737375)
2 = 3079.65557: (1.0377559416905329, 0.026409155737540727, 0.06085124165679545, 7.933610519310132)
3 = 3018.64212: (1.039017565704735, 0.02867894342305815, -0.3677363629374058, 7.768692210518939)
4 = 3003.68458: (1.046396849846413, 0.035961462377881295, -0.3710404168595591, 7.373696771833709)
5 = 2938.55579: (1.0921867088444825, 0.07992511998431093, 0.8597073404976727, 5.188299340115806)
6 = 2529.65909: (1.0927813193235378, 0.08194908415601442, 0.4839814105969523, 5.023471729754752)
7 = 2500.46065: (1.0969523810001807, 0.08862769994997295, 0.29607665009101825, 4.612666685477235)
8 = 2466.73828: (1.14201708029425, 0.15413669464077112, 0.638943702784333, 1.0778375657601904)
9 = 2106.82967: (1.252176088484689, 0.12782680993619894, 52.724672339883604, 17.03988508920688)
10 = 0.00000: (1.252176088484689, 0.12782680993619894, 52.724672339883604, 17.03988508920688)
11 = 0.00000: (1.252176088484689, 0.12782680993619894, 52.724672339883604, 17.03988508920688)
12 = 0.00000: (1.252176088484689, 0.12782680993619894, 52.724672339883604, 17.03988508920688)
13 = 0.00000: (1.252176088484689, 0.12782680993619894, 52.724672339883604, 17.03988508920688)
14 = 0.00000: (1.252176088484689, 0.12782680993619894, 52.724672339883604, 17.03988508920688)
15 = 0.00000: (1.252176088484689, 0.12782680993619894, 52.724672339883604, 17.03988508920688)
16 = 0.00000: (1.252176088484689, 0.12782680993619894, 52.724672339883604, 17.03988508920688)
17 = 0.00000: (1.252176088484689, 0.12782680993619894, 52.724672339883604, 17.03988508920688)
18 = 0.00000: (1.252176088484689, 0.12782680993619894, 52.724672339883604, 17.03988508920688)
In the beginning everything seemed fine. However, something went wrong in iteration 9 and the image was moved out of the boundaries.
Now, setting the Upper Limit to 2.0, it does move the image to the oppsite direction:
0 = 3737.46839: (1.0041671528397889, 0.0018283064652219257, 0.17636473777494874, 1.024814393871803)
1 = 3629.39366: (1.0107770784283137, 0.006177979096732334, 0.07004986540472673, 2.816138032744643)
2 = 3449.24781: (1.0246025518661466, 0.016771881321277125, -0.23374605322512992, 6.8002443859052)
3 = 3075.17899: (1.0403193340035892, 0.030869198918167275, -0.27440774838637527, 8.05680912735824)
4 = 2996.23216: (1.0485623645064288, 0.038808653864649115, -0.4024790191687574, 8.324957828392186)
5 = 2959.73323: (1.066333755973519, 0.05634197231797608, -0.5136948195200061, 8.189624235524011)
6 = 2918.05000: (1.1118821929339107, 0.10167926888790804, 0.020252426908466736, 6.266436226946532)
7 = 2517.51989: (1.158774695166431, 0.15403474685102814, 1.7398572654977367, 0.43751608354740057)
8 = 1754.47276: (1.2631504313988031, 0.2736329845260086, 7.6529750802519105, -5.239059105666371)
WARNING: In /tmp/SimpleITK-build/ITK-prefix/include/ITK-4.13/itkObjectToObjectMetric.hxx, line 529
MeanSquaresImageToImageMetricv4 (0x30e7090): No valid points were found during metric evaluation. For image metrics, verify that the images overlap appropriately. For instance, you can align the image centers by translation. For point-set metrics, verify that the fixed points, once transformed into the virtual domain space, actually lie within the virtual domain.
9 = 935.00137: (1.221492711494796, 0.43658832317571805, -50.477604042183195, -9.305902149954402)
10 = 0.00000: (1.221492711494796, 0.43658832317571805, -50.477604042183195, -9.305902149954402)
11 = 0.00000: (1.221492711494796, 0.43658832317571805, -50.477604042183195, -9.305902149954402)
12 = 0.00000: (1.221492711494796, 0.43658832317571805, -50.477604042183195, -9.305902149954402)
13 = 0.00000: (1.221492711494796, 0.43658832317571805, -50.477604042183195, -9.305902149954402)
14 = 0.00000: (1.221492711494796, 0.43658832317571805, -50.477604042183195, -9.305902149954402)
15 = 0.00000: (1.221492711494796, 0.43658832317571805, -50.477604042183195, -9.305902149954402)
16 = 0.00000: (1.221492711494796, 0.43658832317571805, -50.477604042183195, -9.305902149954402)
17 = 0.00000: (1.221492711494796, 0.43658832317571805, -50.477604042183195, -9.305902149954402)
18 = 0.00000: (1.221492711494796, 0.43658832317571805, -50.477604042183195, -9.305902149954402)
For some reason, I do even get a warning now, that the image is moved out of the image domain.
A few other things I already found out:
- The registration seems to be sensitive to too much empty space in the images. The problem is, I need to register several images and do adjust the image size to the largest in the set. All my images contain a lot of background pixels.
- For some obscure reason, I could make it work in some cases by removing or adding one pixel in the y direction - thus it looks like there is some influence of certain image sizes only
- In some cases, a non-uniform pixel spacing makes the difference. For example when setting the spacing to (0.25, 0.5) it worked, while (1.0, 1.0) did not.
- choosing between Correlation or MeanSquares metric has an influence
- using different interpolators has an influence, however as I’m using usually binary masks, I use NearestNeighbor a lot.
- Removing ScalesFromPhysicalShift and/or replacing with IndexShift has a huge negative effect
It would be interesting to debug this particular case and see why the optimizer behaves so weirdly. But I could not find a way to print out the optimizer’s internal state, i.e. which limits are reached etc.
Is there anything I can do? What would be a solution to register such binary images in a stable manner?
Attached is my code and the two test images.

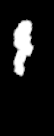
metric_evaluation.py (2.3 KB)
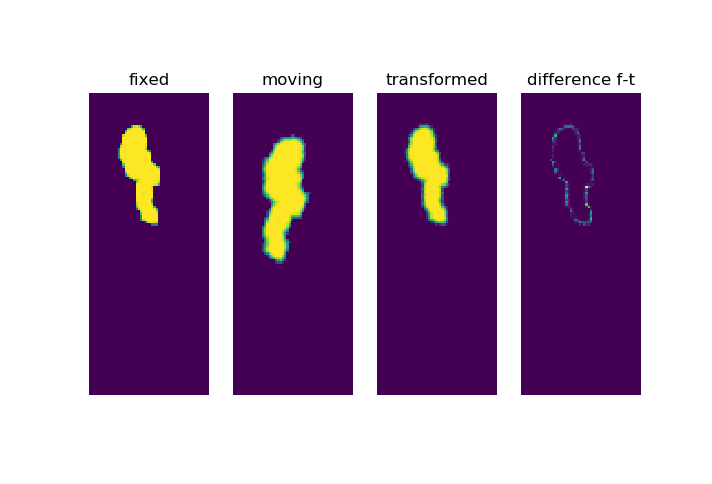
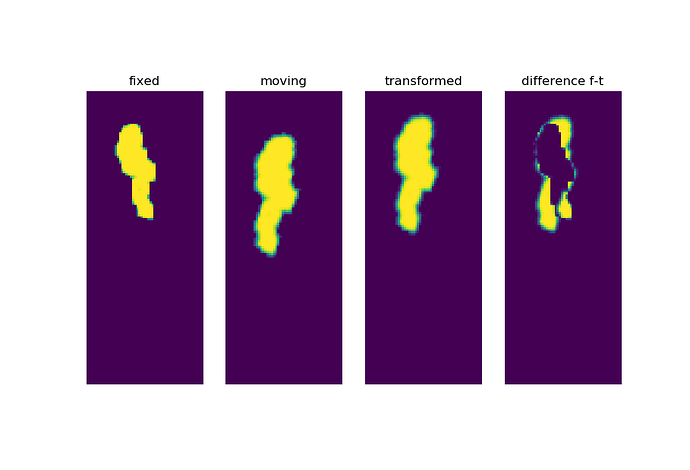
 Thanks for pointing me to the distance maps, I’ll test those!
Thanks for pointing me to the distance maps, I’ll test those!