Hi, I’ve looked for solutions here but i couldnt find. Sorry if it’s a duplication.
I have 11 different DICOM volumes of same patient (converted to nifti) with absolutely different dimensions.
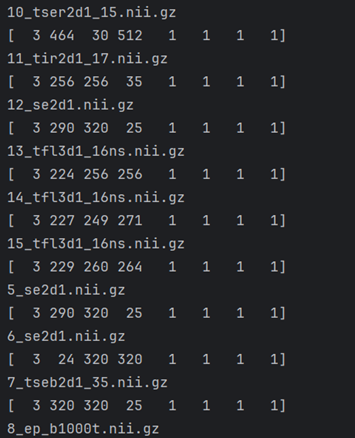
I want to use caPTk for segmentation but caPTk doesnt allow me to use these volumes because of different sizes dims spacing etc.
How can i solve this problem. What is the workflow for this problem?
Thank you.