@zivy ,
Thanks again for the reply. Based on you recommendations, these are the steps I followed:
- Register original images
data_directory = r"venous"
series_IDs = sitk.ImageSeriesReader.GetGDCMSeriesIDs(data_directory)
series_file_names = sitk.ImageSeriesReader.GetGDCMSeriesFileNames(data_directory, series_IDs[0])
data_directory2 = r"arterial"
series_IDs2 = sitk.ImageSeriesReader.GetGDCMSeriesIDs(data_directory2)
series_file_names2 = sitk.ImageSeriesReader.GetGDCMSeriesFileNames(data_directory2, series_IDs2[0])
fixed_image = sitk.ReadImage(series_file_names, sitk.sitkFloat32)
moving_image = sitk.ReadImage(series_file_names2, sitk.sitkFloat32)
initial_transform = sitk.CenteredTransformInitializer(fixed_image,
moving_image,
sitk.ComposeScaleSkewVersor3DTransform(),
sitk.CenteredTransformInitializerFilter.GEOMETRY)
moving_resampled = sitk.Resample(moving_image, fixed_image, initial_transform, sitk.sitkLinear, -1000, moving_image.GetPixelID())
registration_method = sitk.ImageRegistrationMethod()
# Similarity metric settings.
registration_method.SetMetricAsMattesMutualInformation(numberOfHistogramBins=50)
registration_method.SetMetricSamplingStrategy(registration_method.RANDOM)
registration_method.SetMetricSamplingPercentage(0.01)
registration_method.SetInterpolator(sitk.sitkLinear)
# Optimizer settings.
registration_method.SetOptimizerAsGradientDescent(learningRate=1.0, numberOfIterations=100, convergenceMinimumValue=1e-6,
convergenceWindowSize=10)
registration_method.SetOptimizerScalesFromPhysicalShift()
# Don't optimize in-place, we would possibly like to run this cell multiple times.
registration_method.SetInitialTransform(initial_transform, inPlace=False)
# Connect all of the observers so that we can perform plotting during registration.
registration_method.AddCommand(sitk.sitkStartEvent, start_plot)
registration_method.AddCommand(sitk.sitkEndEvent, end_plot)
registration_method.AddCommand(sitk.sitkMultiResolutionIterationEvent, update_multires_iterations)
registration_method.AddCommand(sitk.sitkIterationEvent, lambda: plot_values(registration_method))
final_transform = registration_method.Execute(sitk.Cast(fixed_image, sitk.sitkFloat32),
sitk.Cast(moving_image, sitk.sitkFloat32))
print(f'Final metric value: {registration_method.GetMetricValue()}')
print(f'Optimizer\'s stopping condition, {registration_method.GetOptimizerStopConditionDescription()}')
moving_resampled = sitk.Resample(moving_image, fixed_image, final_transform, sitk.sitkLinear, 0.0, moving_image.GetPixelID())
interact(display_images, fixed_image_z=(0,fixed_image.GetSize()[2]-1), moving_image_z=(0,moving_image.GetSize()[2]-1),
fixed_npa = fixed(sitk.GetArrayViewFromImage(fixed_image)), moving_npa=fixed(sitk.GetArrayViewFromImage(moving_image)));
interact(display_images_with_alpha, image_z=(0,fixed_image.GetSize()[2] - 1), alpha=(0.0,1.0,0.05),
fixed = fixed(fixed_image), moving=fixed(moving_resampled));
After registration, the two images are aligned and have same domain: size, spacing, orgin, direction.
(512, 512, 156)
(0.869140625, 0.869140625, 3.0)
(-231.0654296875, -391.0654296875, 37.5)
(1.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 1.0)
- Next I transformed the fixed image so that it has isotropic voxel values:
def resample_image(image, out_spacing=(1.0, 1.0, 1.0), out_size=None, is_label=False, pad_value=0):
"""Resamples an image to given element spacing and output size."""
original_spacing = np.array(image.GetSpacing())
original_size = np.array(image.GetSize())
if out_size is None:
out_size = np.round(np.array(original_size * original_spacing / np.array(out_spacing))).astype(int)
else:
out_size = np.array(out_size)
original_direction = np.array(image.GetDirection()).reshape(len(original_spacing),-1)
original_center = (np.array(original_size, dtype=float) - 1.0) / 2.0 * original_spacing
out_center = (np.array(out_size, dtype=float) - 1.0) / 2.0 * np.array(out_spacing)
original_center = np.matmul(original_direction, original_center)
out_center = np.matmul(original_direction, out_center)
out_origin = np.array(image.GetOrigin()) + (original_center - out_center)
resample = sitk.ResampleImageFilter()
resample.SetOutputSpacing(out_spacing)
resample.SetSize(out_size.tolist())
resample.SetOutputDirection(image.GetDirection())
resample.SetOutputOrigin(out_origin.tolist())
resample.SetTransform(sitk.Transform())
resample.SetDefaultPixelValue(pad_value)
if is_label:
resample.SetInterpolator(sitk.sitkNearestNeighbor)
else:
#resample.SetInterpolator(sitk.sitkBSpline)
resample.SetInterpolator(sitk.sitkLinear)
return resample.Execute(sitk.Cast(image, sitk.sitkFloat32))
Now the image domain is as follows:
(445, 445, 468)
(1.0, 1.0, 1.0)
(-231.0, -391.0, 36.5)
(1.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 1.0)
- I then resampled the moving_resampled image (obtained after registration) to isotopic fixed image domain:
resnew = sitk.Resample(moving_resampled, isotropic_image)
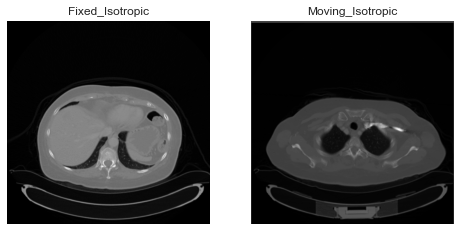
While this image has same domain as the isotropic image, slices do not overlap. e.g: slice 100 in isotropic fixed image is different than that of slice 100 in resampled moving image onto istropic image.
However, if I do sitk.Resample(moving_image, isotropic_image) they do overlap.
What am I doing wrong here? Why does slices do not overlap after registration even if I resample to isotropic_image. ?