I have a binary image stack or space of binarized voxels (either black or white voxels).
Since my data is from electron microscopy the spacing of the pixels are (1, 1, Z) where (x, y, z), and Z is an arbitrary value defined by the limitation of preparatory steps before performing the imaging with the electron microscope.
My current issue is resampling. With this code:
def resample(binary_stack: np.ndarray, z_spacing: float) -> np.ndarray:
binary_stack_itk = sitk.GetImageFromArray(binary_stack.astype(np.float32))
new_size = list(binary_stack_itk.GetSize())
new_size[2] = round(z_spacing * new_size[2])
return sitk.GetArrayFromImage(sitk.Resample(
binary_stack_itk,
new_size,
sitk.Transform(),
sitk.sitkLinear,
binary_stack_itk.GetOrigin(),
(1.0, 1.0, 1.0 / z_spacing),
binary_stack_itk.GetDirection(),
0,
binary_stack_itk.GetPixelID(),
)).astype(bool)
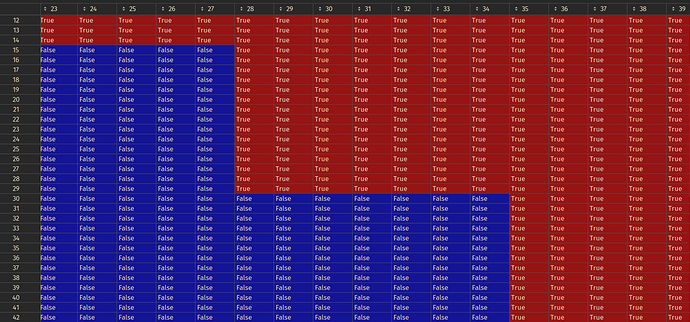
If I rotate the mesh 90 degrees (np.rot90(result)) we can inspect the array along the zy-plane. The following is an arbitrary position of somewhere in the middle of the zy-plane in one of my resampled binary stacks:
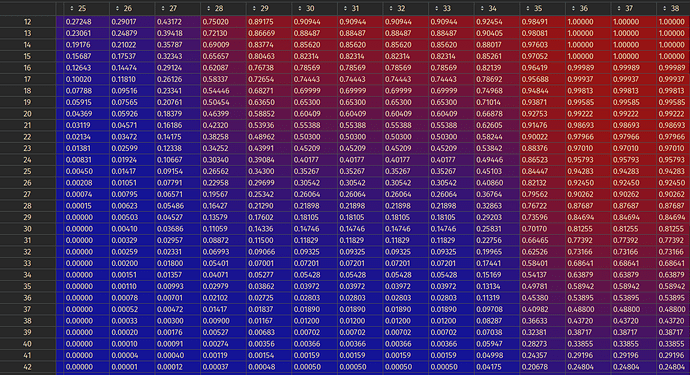
This is not smooth at all! I tried changing methods from sitk.sitkLinear sitk.sitkBSplineResamplerOrder3, and get this when rotated:
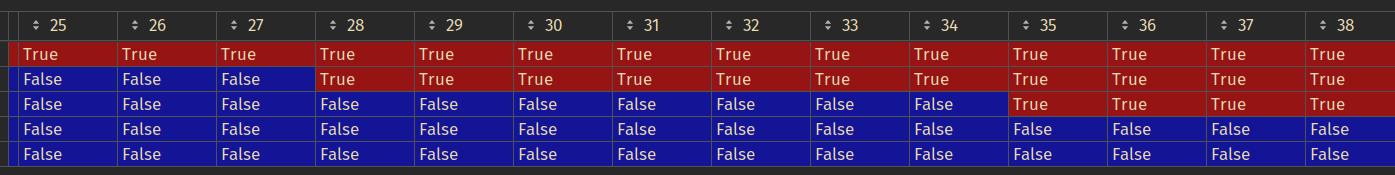
Before resample, segmantation masks stacked on top of each other (same location as the ones above), also rotated (np.rot90(stack)):
The resample is way too simple with respect to what I am looking for. I was hoping to find a resample that would be smoother, natively. I would like the resample to be connecting the edges of the segmentations along zy plane so that they are connected along their edges, not something like the one we see in sitkLinear, where the previous edge is traced to the next edge:
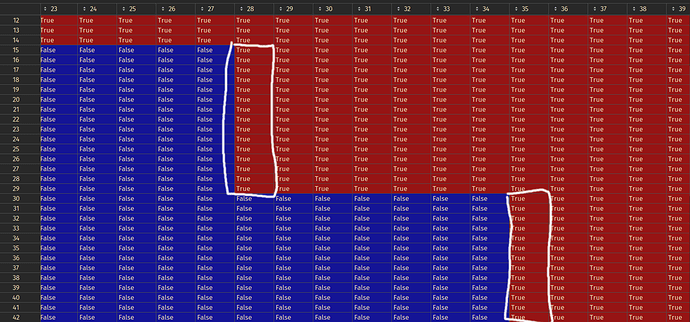
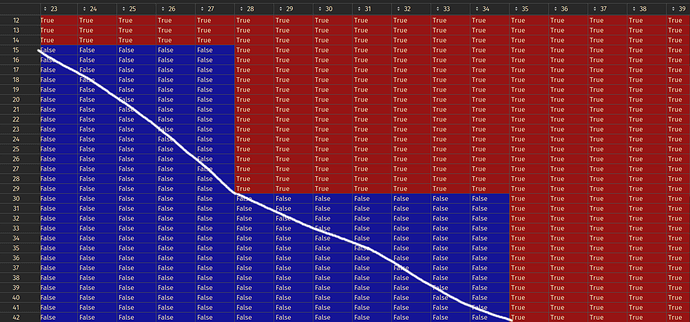
I would like something like this instead:
Where everything above the white line is True!
Any resample method that I missed in the Simple ITK liberary, or is what I am looking for non-existant? I have to avoid functions that would require variable tuning of any sort as these introduce bias to the data.