Hi all,
Long-time reader, first-time poster here! I’m not sure if this is the right category, but as it’s my first question, I’ll post it here.
I am performing deformable image registration using DEMONS in a 3D domain between same-modality medical images. In the registration process, I encounter large variances in the sizes of the structures I need to register—some are large, while others are very small. Every structure is equally important for this task, and if some are registered well while others are less accurate, the overall result is suboptimal for my needs.
One advantage I have is a rough mask of where the movement is occurring. I’ve tried using itk::simple::ImageRegistrationMethod::SetMetricFixedMask to evaluate the registration only in those regions, but this yielded very little improvement.
I’ve also experimented with:
- Various combinations of iteration numbers, smoothing sigmas, convergence values, etc.
- All available optimizers for DEMONS,
- Different pyramidal registration strategies,
- Multiscale DEMONS using an initial affine registration followed by DEMONS.
Current Approach and Issue
Now, I’m exploring the idea of obtaining a localized motion vector field by subdividing the 3D volume into smaller, non-uniform 3D subvolumes based on prior knowledge of structure locations and movement ranges.
However, the main challenge I face is how to merge these localized motion vector fields of varying sizes into a single cohesive field. The worflow I was planning to implement is as follows:
- Original Image: Size
(N, M, K)with spacing(s, s, s)(equal spacing). - Subvolumes: Divide the image into multiple subvolumes with sizes
(Ni, Mi, Ki)such that:Sum(Ni) = N,Sum(Mi) = M,Sum(Ki) = K(fori = 0, ..., n).
- DEMONS Registration: Perform DEMONS registration on each subvolume.
- Merge Motion Fields: Combine the subvolume motion vector fields to apply them to the original image of size
(N, M, K).
One workaround I’m considering is:
- Converting the motion vector fields into NumPy arrays of size
(Ni, Mi, Ki, 3)for each subvolume, - Merging these arrays into a single array of size
(N, M, K, 3), and - Returning to the SimpleITK framework to apply the deformation field.
However, this approach feels very suboptimal, as I’ve never switched between NumPy and the ITK/SimpleITK framework for this type of task before.
Questions
- Is there a better way to perform step 3 (DEMONS registration on subvolumes)?
- Is there a more appropriate way to achieve my overall goal (e.g., using another type of weighting or mask maybe)?
- Has anyone faced a similar issue? Any leads on this would be greatly appreciated!
Thanks a lot for your help!
P.S.:
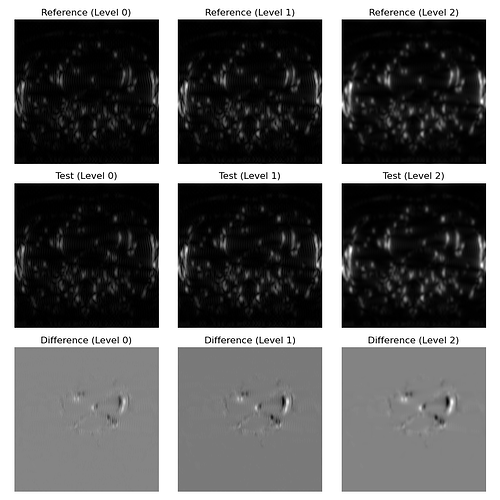
To add context, these are the images I am trying to register in a pyramidal fashion. Top row is the fixed image, middle row is the moving image, bottom row is the difference between movbing and fixed:
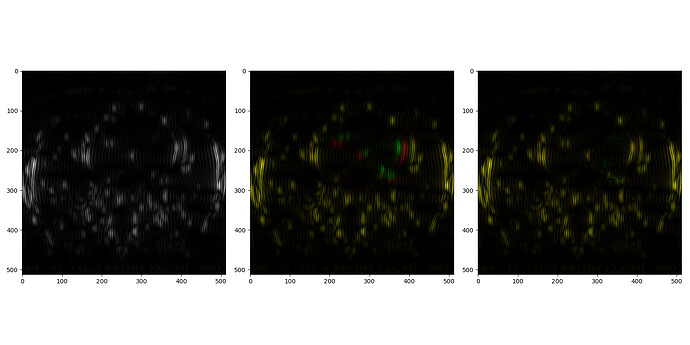
In 2D I can register all the structures very well (see image), but in 3D it has been almost impossible to get the right motion vector field for the small structure on the upper left structure of the fixed-moving image.