I have two dicom series images of brain. I want to perform registration between the images in ITK. I wanted to find deformation points first, do transformation, then apply inverse deformation. Suggest me a suitable algorithm available in ITK
The examples you could take a look at for inspiration are Read DICOM Series and BSpline registration.
I coudn’t find CMakeLists.txt file here. Can you please help me?
Here are some instructions on building Sphinx examples. WikiExamples have a CMakeLists.txt on each page.
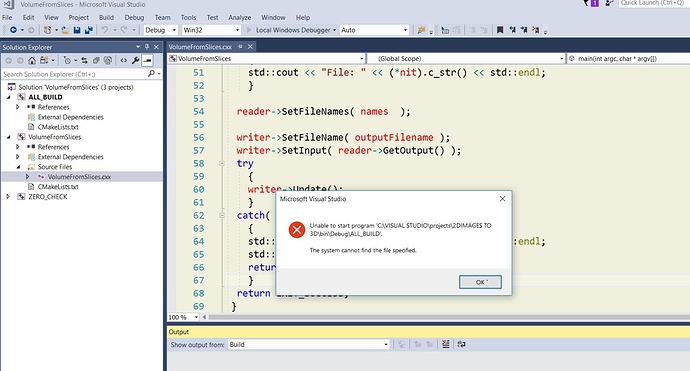
thank you…Can you guide me on how to debug this codes in visual studio? I have my .cxx file and CMakeLists.txt file in source folder and created a bin folder for building. I am able to build this code in visual studio. I want to learn and understand the code line by line by using the debugger. But it is throwing an error as below
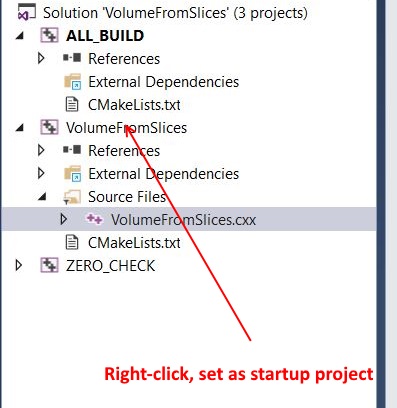
You should right click on VolumeFromSlices, and choose option Set as StartUp project.
Thank you so much sir. I really didnt expect such a fast reply . Where can i some more details on debugging this code? any tutorial or document? i am completely new to this.
You should look at Microsoft’s documentation.
Additionally, natvis files enable more useful debug views of objects. We have started writing natvis file for ITK.