Hello,
I am new to image registration and I want to register an MRI Head Image with a CT image.
I began with the example from this Topic to understand how image registration is working in SITK:
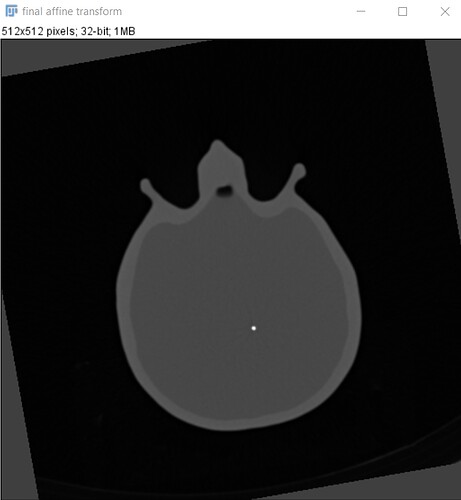
But when I applied the code on my MRI and CT images the result look like the input image.
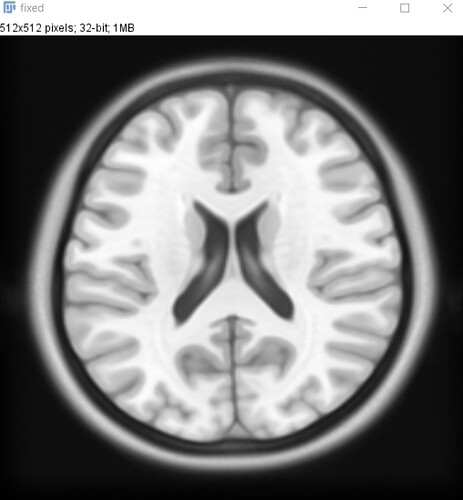
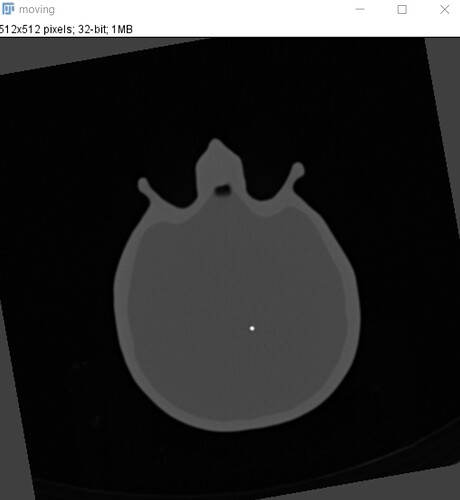
As an input(fixed image) I chose the MRI image and the moving image is the CT image. Both images have the same size(512x512).
Here is my fixed image:
Here ist the moving image:
And here is the result:
def multires_registration(fixed_image, moving_image, initial_transform):
registration_method = sitk.ImageRegistrationMethod()
registration_method.SetInterpolator(sitk.sitkBSpline)
registration_method.SetMetricAsMattesMutualInformation(numberOfHistogramBins=50)
registration_method.SetMetricSamplingStrategy(registration_method.RANDOM)
registration_method.SetMetricSamplingPercentage(0.01)
registration_method.SetOptimizerAsGradientDescent(learningRate=0.5, numberOfIterations=100, convergenceMinimumValue=1e-6, convergenceWindowSize=10)
registration_method.SetOptimizerScalesFromPhysicalShift()
registration_method.SetShrinkFactorsPerLevel(shrinkFactors = [2,1])
registration_method.SetSmoothingSigmasPerLevel(smoothingSigmas = [1,0])
registration_method.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
optimized_transform = sitk.AffineTransform(2)
registration_method.SetMovingInitialTransform(initial_transform)
registration_method.SetInitialTransform(optimized_transform, inPlace=False)
#registration_method.AddCommand(sitk.sitkStartEvent, registration_callbacks.metric_start_plot)
#registration_method.AddCommand(sitk.sitkEndEvent, registration_callbacks.metric_end_plot)
#registration_method.AddCommand(sitk.sitkMultiResolutionIterationEvent, registration_callbacks.metric_update_multires_iterations)
#registration_method.AddCommand(sitk.sitkIterationEvent, lambda: registration_callbacks.metric_plot_values(registration_method))
optimized_transform = registration_method.Execute(fixed_image, moving_image)
print('Final metric value: {0}'.format(registration_method.GetMetricValue()))
print('Optimizer\'s stopping condition, {0}'.format(registration_method.GetOptimizerStopConditionDescription()))
print('Optimized Transform from Multires:')
print(optimized_transform)
return (optimized_transform)
sitk.Show(resample_Atlas_Image2, 'fixed')
sitk.Show(phantom_image2, 'moving')
sitk.Show(sitk.Resample(phantom_image2, resample_Atlas_Image2, sitk.Transform()), 'identity transform')
# Centered 2D affine transform and show the resampled moving_image using this transform.
registration_transform = sitk.CenteredTransformInitializer(resample_Atlas_Image2,
phantom_image2,
sitk.AffineTransform(2),
sitk.CenteredTransformInitializerFilter.GEOMETRY)
sitk.Show(sitk.Resample(phantom_image2, resample_Atlas_Image2, registration_transform), 'initial affine transform')
# Register using 2D affine initial transform that is overwritten
# and show the resampled moving_image using this transform.
multires_registration(resample_Atlas_Image2, phantom_image2, registration_transform)
sitk.Show(sitk.Resample(phantom_image2, resample_Atlas_Image2, registration_transform), 'final affine transform')
Could anybody help me with this?
Thank you very much!