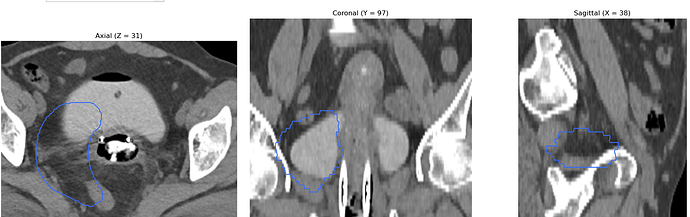
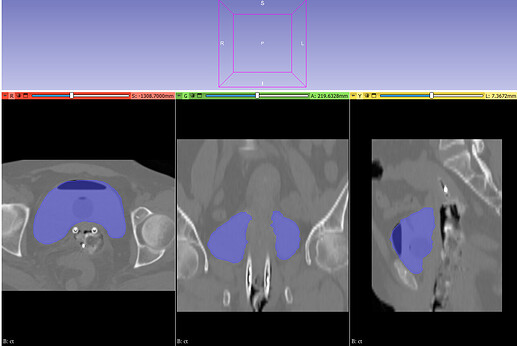
Hello everyone,
I am working on displaying CT images in the axial, coronal, and sagittal planes, along with dose maps and RTSTRUCT contours. While the CT and dose images are displayed correctly, I am having trouble overlaying the structures (contours) from the RTSTRUCT file on the CT images—the contours do not align properly with the CT, as shown in the attached image.
What steps should I take to ensure the contours are properly overlaid on the CT images in all three planes? Can you help me with this issue?
thanks a lot
my code:
import pydicom
import numpy as np
import matplotlib.pyplot as plt
import ipywidgets as widgets
from ipywidgets import interact, HBox, VBox, Output
import os
import SimpleITK as sitk
from rt_utils import RTStructBuilder
from scipy.ndimage import zoom
from skimage.measure import find_contours
import warnings
warnings.filterwarnings('ignore')
class MedicalImageViewer:
def __init__(self, ct_path, dose_paths=None, rtstruct_path=None):
"""
Initialize the medical image viewer.
"""
self.ct_path = ct_path
self.dose_paths = dose_paths or {}
self.rtstruct_path = rtstruct_path
# Data storage
self.ct_volume = None
self.ct_metadata = None
self.dose_volumes = {}
self.contours = {}
self.roi_names = []
# Display parameters
self.window_level = (40, 400)
self.dose_colormap = plt.cm.jet
self.dose_alpha = 0.4
# Orientation
self.patient_position = 'HFS'
self.load_all_data()
def load_ct(self):
"""Load CT volume."""
print("Loading CT images...")
reader = sitk.ImageSeriesReader()
dicom_names = reader.GetGDCMSeriesFileNames(self.ct_path)
if not dicom_names: raise ValueError("No DICOM files found in CT path")
reader.SetFileNames(dicom_names)
ct_image = reader.Execute()
array = sitk.GetArrayFromImage(ct_image)
self.ct_volume = np.transpose(array, (1, 2, 0))
first_slice = pydicom.dcmread(dicom_names[0])
self.patient_position = first_slice.PatientPosition if hasattr(first_slice, 'PatientPosition') else 'HFS'
self.ct_metadata = {
'pixel_spacing': first_slice.PixelSpacing if hasattr(first_slice, 'PixelSpacing') else [1.0, 1.0],
'slice_thickness': float(first_slice.SliceThickness) if hasattr(first_slice, 'SliceThickness') else 1.0
}
print(f"CT loaded: {self.ct_volume.shape}")
def detect_sagittal_flip(self):
"""Detect if sagittal flip is needed."""
return 'FF' in self.patient_position.upper()
def load_doses(self):
"""Load dose volumes."""
for dose_name, dose_path in self.dose_paths.items():
print(f"Loading {dose_name} dose...")
ds = pydicom.dcmread(dose_path)
dose_array = ds.pixel_array.astype(np.float32)
dose_volume = dose_array * float(ds.DoseGridScaling) if hasattr(ds, 'DoseGridScaling') else dose_array
dose_volume = np.transpose(dose_volume, (1, 2, 0))
if dose_volume.shape != self.ct_volume.shape:
zoom_factors = [self.ct_volume.shape[i] / dose_volume.shape[i] for i in range(3)]
dose_volume = zoom(dose_volume, zoom_factors, order=1)
self.dose_volumes[dose_name] = dose_volume
print(f"{dose_name} dose loaded: {dose_volume.shape}")
def load_rtstruct(self):
"""Load RTSTRUCT masks."""
if not self.rtstruct_path or not os.path.exists(self.rtstruct_path): return
print("Loading RTSTRUCT...")
rtstruct = RTStructBuilder.create_from(dicom_series_path=self.ct_path, rt_struct_path=self.rtstruct_path)
self.roi_names = rtstruct.get_roi_names()
colors = plt.cm.rainbow(np.linspace(0, 1, len(self.roi_names)))
for i, roi_name in enumerate(self.roi_names):
mask = rtstruct.get_roi_mask_by_name(roi_name)
mask = np.transpose(mask, (1, 0, 2))
self.contours[roi_name] = {'mask': mask, 'color': colors[i]}
print(f"Loaded {len(self.contours)} ROIs")
def load_all_data(self):
"""Load all data."""
self.load_ct()
self.load_doses()
self.load_rtstruct()
def apply_window_level(self, image, window, level):
"""Apply window/level to CT."""
img_min = level - window / 2
img_max = level + window / 2
windowed = np.clip(image, img_min, img_max)
return (windowed - img_min) / (img_max - img_min)
def create_interactive_viewer(self):
"""Create the fully interactive viewe."""
ct_shape = self.ct_volume.shape
pixel_spacing = self.ct_metadata['pixel_spacing']
slice_thickness = self.ct_metadata['slice_thickness']
axial_aspect = pixel_spacing[1] / pixel_spacing[0]
sagittal_aspect = slice_thickness / pixel_spacing[1]
coronal_aspect = slice_thickness / pixel_spacing[0]
sagittal_flip = self.detect_sagittal_flip()
output = Output()
# --- Sliders

---
axial_slider = widgets.IntSlider(min=0, max=ct_shape[2]-1, value=ct_shape[2]//2, description='Axial (Z):', continuous_update=False)
coronal_slider = widgets.IntSlider(min=0, max=ct_shape[0]-1, value=ct_shape[0]//2, description='Coronal (Y):', continuous_update=False)
sagittal_slider = widgets.IntSlider(min=0, max=ct_shape[1]-1, value=ct_shape[1]//2, description='Sagittal (X):', continuous_update=False)
window_slider = widgets.IntSlider(min=1, max=2000, value=self.window_level[1], description='Window:')
level_slider = widgets.IntSlider(min=-1000, max=1000, value=self.window_level[0], description='Level:')
dose_selector = widgets.Dropdown(options=['None'] + list(self.dose_volumes.keys()), value='None', description='Dose:')
dose_alpha_slider = widgets.FloatSlider(min=0, max=1, value=self.dose_alpha, step=0.1, description='Opacity:')
roi_selector = widgets.SelectMultiple(options=self.roi_names, value=[], description='ROIs:', rows=5)
def update_display(axial_idx, coronal_idx, sagittal_idx, window, level, dose_name, dose_alpha, selected_rois):
with output:
output.clear_output(wait=True)
fig, axes = plt.subplots(1, 3, figsize=(18, 6))
ax_axial, ax_coronal, ax_sagittal = axes # Correct plot order
# --- Axial View ---
axial_ct = np.flipud(self.ct_volume[:, :, axial_idx])
axial_ct = self.apply_window_level(axial_ct, window, level)
ax_axial.imshow(axial_ct, cmap='gray', aspect=axial_aspect, origin='lower')
ax_axial.set_title(f'Axial (Z = {axial_idx})')
# --- Coronal View ---
coronal_ct = np.flipud(self.ct_volume[coronal_idx, :, :].T[::-1])
coronal_ct = self.apply_window_level(coronal_ct, window, level)
ax_coronal.imshow(coronal_ct, cmap='gray', aspect=coronal_aspect, origin='lower')
ax_coronal.set_title(f'Coronal (Y = {coronal_idx})')
# --- Sagittal View ---
sagittal_ct = self.ct_volume[:, sagittal_idx, :].T
if sagittal_flip: sagittal_ct = np.flipud(sagittal_ct)
else: sagittal_ct = np.fliplr(sagittal_ct)
sagittal_ct = self.apply_window_level(sagittal_ct, window, level)
ax_sagittal.imshow(sagittal_ct, cmap='gray', aspect=sagittal_aspect, origin='lower')
ax_sagittal.set_title(f'Sagittal (X = {sagittal_idx})')
# --- Dose Overlay ---
if dose_name != 'None' and dose_name in self.dose_volumes:
dose_vol = self.dose_volumes[dose_name]
max_dose = np.max(dose_vol)
dose_axial = np.flipud(dose_vol[:, :, axial_idx])
im = ax_axial.imshow(np.ma.masked_equal(dose_axial, 0), cmap=self.dose_colormap, alpha=dose_alpha, vmin=0, vmax=max_dose, aspect=axial_aspect, origin='lower')
dose_coronal = np.flipud(dose_vol[coronal_idx, :, :].T[::-1])
ax_coronal.imshow(np.ma.masked_equal(dose_coronal, 0), cmap=self.dose_colormap, alpha=dose_alpha, vmin=0, vmax=max_dose, aspect=coronal_aspect, origin='lower')
dose_sagittal = dose_vol[:, sagittal_idx, :].T
if sagittal_flip: dose_sagittal = np.flipud(dose_sagittal)
else: dose_sagittal = np.fliplr(dose_sagittal)
ax_sagittal.imshow(np.ma.masked_equal(dose_sagittal, 0), cmap=self.dose_colormap, alpha=dose_alpha, vmin=0, vmax=max_dose, aspect=sagittal_aspect, origin='lower')
plt.colorbar(im, ax=axes, fraction=0.046, pad=0.04, label='Dose (GY)')
# --- Structure Overlay ---
for roi_name in selected_rois:
if roi_name in self.contours:
mask = self.contours[roi_name]['mask']
color = self.contours[roi_name]['color']
# Axial Structure
axial_mask = np.flipud(mask[:, :, axial_idx])
contours_found = find_contours(axial_mask, 0.5)
for c in contours_found: ax_axial.plot(c[:, 1], c[:, 0], color=color, linewidth=1.5)
# Coronal Structure
coronal_mask = np.flipud(mask[coronal_idx, :, :].T[::-1])
contours_found = find_contours(coronal_mask, 0.5)
for c in contours_found: ax_coronal.plot(c[:, 1], c[:, 0], color=color, linewidth=1.5)
# Sagittal Structure
sagittal_mask = mask[:, sagittal_idx, :].T
if sagittal_flip: sagittal_mask = np.flipud(sagittal_mask)
else: sagittal_mask = np.fliplr(sagittal_mask)
contours_found = find_contours(sagittal_mask, 0.5)
for c in contours_found: ax_sagittal.plot(c[:, 1], c[:, 0], color=color, linewidth=1.5)
for ax in axes: ax.axis('off')
plt.tight_layout()
plt.show()
# --- Linking sliders to the correct function arguments ---
interact(update_display, axial_idx=axial_slider, coronal_idx=coronal_slider, sagittal_idx=sagittal_slider,
window=window_slider, level=level_slider, dose_name=dose_selector, dose_alpha=dose_alpha_slider, selected_rois=roi_selector)
display(output)
# directories
if __name__ == "__main__":
ct_path = r'D:\TEST\CT'
dose_paths = {
'Physical': r'D:\TEST\RTDOSE\RTDOSE_Dose.dcm',
}
rtstruct_path = r'D:\TEST\RTSTRUCT\RTSTRUCT.dcm'
viewer = MedicalImageViewer(ct_path=ct_path, dose_paths=dose_paths, rtstruct_path=rtstruct_path)
viewer.create_interactive_viewer()