I am trying to register the POPI model using BSpline Transform (Code given in SimpleITK Tutorial - 06_advanced_registration.ipynb). However instead of TRE used in the Tutorial, I am trying to compute other accuracy metrices like Jaccard coefficient, Dice coefficient and HausdorfDistance. The entire code is given below
import SimpleITK as sitk
import registration_gui as rgui
import utilities
from downloaddata import fetch_data as fdata
from ipywidgets import interact, fixed
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
images = []
masks = []
points = []
image_indexes = [0,7]
for i in image_indexes:
image_file_name = 'POPI/meta/{0}0-P.mhd'.format(i)
mask_file_name = 'POPI/masks/{0}0-air-body-lungs.mhd'.format(i)
points_file_name = 'POPI/landmarks/{0}0-Landmarks.pts'.format(i)
images.append(sitk.ReadImage(fdata(image_file_name), sitk.sitkFloat32))
masks.append(sitk.ReadImage(fdata(mask_file_name)))
points.append(utilities.read_POPI_points(fdata(points_file_name)))
interact(rgui.display_coronal_with_overlay, temporal_slice=(0,len(images)-1),
coronal_slice = (0, images[0].GetSize()[1]-1),
images = fixed(images), masks = fixed(masks),
label= fixed(utilities.popi_lung_label), window_min = fixed(-1024), window_max=fixed(976));
fixed_index = 0
moving_index = 1
fixed_image = images[fixed_index]
fixed_image_mask = masks[fixed_index] == utilities.popi_lung_label
fixed_points = points[fixed_index]
moving_image = images[moving_index]
moving_image_mask = masks[moving_index] == utilities.popi_lung_label
moving_points = points[moving_index]
# Define a simple callback which allows us to monitor registration progress.
def iteration_callback(filter):
print('\r{0:.2f}'.format(filter.GetMetricValue()), end='')
registration_method = sitk.ImageRegistrationMethod()
# Determine the number of BSpline control points using the physical
# spacing we want for the finest resolution control grid.
grid_physical_spacing = [50.0, 50.0, 50.0] # A control point every 50mm
image_physical_size = [size*spacing for size,spacing in zip(fixed_image.GetSize(),
fixed_image.GetSpacing())]
mesh_size = [int(image_size/grid_spacing + 0.5) \
for image_size,grid_spacing in zip(image_physical_size,grid_physical_spacing)]
# The starting mesh size will be 1/4 of the original, it will be refined by
# the multi-resolution framework.
mesh_size = [int(sz/4 + 0.5) for sz in mesh_size]
initial_transform = sitk.BSplineTransformInitializer(image1 = fixed_image,
transformDomainMeshSize = mesh_size, order=3)
# Instead of the standard SetInitialTransform we use the BSpline specific method which also
# accepts the scaleFactors parameter to refine the BSpline mesh. In this case we start with
# the given mesh_size at the highest pyramid level then we double it in the next lower level and
# in the full resolution image we use a mesh that is four times the original size.
registration_method.SetInitialTransformAsBSpline(initial_transform,
inPlace=False,
scaleFactors=[1,2,4])
registration_method.SetMetricAsMeanSquares()
registration_method.SetMetricSamplingStrategy(registration_method.RANDOM)
registration_method.SetMetricSamplingPercentage(0.01)
registration_method.SetMetricFixedMask(fixed_image_mask)
registration_method.SetShrinkFactorsPerLevel(shrinkFactors = [4,2,1])
registration_method.SetSmoothingSigmasPerLevel(smoothingSigmas=[2,1,0])
registration_method.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
registration_method.SetInterpolator(sitk.sitkLinear)
registration_method.SetOptimizerAsLBFGS2(solutionAccuracy=1e-2, numberOfIterations=100, deltaConvergenceTolerance=0.01)
registration_method.AddCommand(sitk.sitkIterationEvent, lambda: iteration_callback(registration_method))
final_transformation = registration_method.Execute(fixed_image, moving_image)
print('\nOptimizer\'s stopping condition, {0}'.format(registration_method.GetOptimizerStopConditionDescription()))
transformed_labels = sitk.Resample(moving_image_mask,
fixed_image,
final_transformation,
sitk.sitkNearestNeighbor,
0.0,
moving_image_mask.GetPixelID())
# Often referred to as ground truth, but we prefer reference as the truth is never known.
reference_segmentation = fixed_image_mask
# Segmentations before and after registration
segmentations = [moving_image_mask, transformed_labels == utilities.popi_lung_label]
from enum import Enum
# Use enumerations to represent the various evaluation measures
class OverlapMeasures(Enum):
jaccard, dice, volume_similarity, false_negative, false_positive = range(5)
class SurfaceDistanceMeasures(Enum):
hausdorff_distance, mean_surface_distance, median_surface_distance, std_surface_distance, max_surface_distance = range(5)
# Empty numpy arrays to hold the results
overlap_results = np.zeros((len(segmentations),len(OverlapMeasures.__members__.items())))
surface_distance_results = np.zeros((len(segmentations),len(SurfaceDistanceMeasures.__members__.items())))
# Compute the evaluation criteria
# Note that for the overlap measures filter, because we are dealing with a single label we
# use the combined, all labels, evaluation measures without passing a specific label to the methods.
overlap_measures_filter = sitk.LabelOverlapMeasuresImageFilter()
hausdorff_distance_filter = sitk.HausdorffDistanceImageFilter()
# Use the absolute values of the distance map to compute the surface distances (distance map sign, outside or inside
# relationship, is irrelevant)
label = 1
reference_distance_map = sitk.Abs(sitk.SignedMaurerDistanceMap(reference_segmentation, squaredDistance=False))
reference_surface = sitk.LabelContour(reference_segmentation)
statistics_image_filter = sitk.StatisticsImageFilter()
# Get the number of pixels in the reference surface by counting all pixels that are 1.
statistics_image_filter.Execute(reference_surface)
num_reference_surface_pixels = int(statistics_image_filter.GetSum())
for i, seg in enumerate(segmentations):
# Overlap measures
overlap_measures_filter.Execute(reference_segmentation, seg)
overlap_results[i,OverlapMeasures.jaccard.value] =
overlap_measures_filter.GetJaccardCoefficient()
overlap_results[i,OverlapMeasures.dice.value] =
overlap_measures_filter.GetDiceCoefficient()
overlap_results[i,OverlapMeasures.volume_similarity.value] =
overlap_measures_filter.GetVolumeSimilarity()
overlap_results[i,OverlapMeasures.false_negative.value] =
overlap_measures_filter.GetFalseNegativeError()
overlap_results[i,OverlapMeasures.false_positive.value] =
overlap_measures_filter.GetFalsePositiveError()
# Hausdorff distance
hausdorff_distance_filter.Execute(reference_segmentation, seg)
surface_distance_results[i,SurfaceDistanceMeasures.hausdorff_distance.value] =
hausdorff_distance_filter.GetHausdorffDistance()
# Symmetric surface distance measures
segmented_distance_map = sitk.Abs(sitk.SignedMaurerDistanceMap(seg,
squaredDistance=False))
segmented_surface = sitk.LabelContour(seg)
# Multiply the binary surface segmentations with the distance maps. The resulting distance
# maps contain non-zero values only on the surface (they can also contain zero on the surface)
seg2ref_distance_map = reference_distance_map*sitk.Cast(segmented_surface, sitk.sitkFloat32)
ref2seg_distance_map = segmented_distance_map*sitk.Cast(reference_surface, sitk.sitkFloat32)
# Get the number of pixels in the segmented surface by counting all pixels that are 1.
statistics_image_filter.Execute(segmented_surface)
num_segmented_surface_pixels = int(statistics_image_filter.GetSum())
# Get all non-zero distances and then add zero distances if required.
seg2ref_distance_map_arr = sitk.GetArrayViewFromImage(seg2ref_distance_map)
seg2ref_distances = list(seg2ref_distance_map_arr[seg2ref_distance_map_arr!=0])
seg2ref_distances = seg2ref_distances + \
list(np.zeros(num_segmented_surface_pixels - len(seg2ref_distances)))
ref2seg_distance_map_arr = sitk.GetArrayViewFromImage(ref2seg_distance_map)
ref2seg_distances = list(ref2seg_distance_map_arr[ref2seg_distance_map_arr!=0])
ref2seg_distances = ref2seg_distances + \
list(np.zeros(num_reference_surface_pixels - len(ref2seg_distances)))
all_surface_distances = seg2ref_distances + ref2seg_distances
surface_distance_results[i,SurfaceDistanceMeasures.mean_surface_distance.value] = np.mean(all_surface_distances)
surface_distance_results[i,SurfaceDistanceMeasures.median_surface_distance.value] = np.median(all_surface_distances)
surface_distance_results[i,SurfaceDistanceMeasures.std_surface_distance.value] = np.std(all_surface_distances)
surface_distance_results[i,SurfaceDistanceMeasures.max_surface_distance.value] = np.max(all_surface_distances)
import pandas as pd
from IPython.display import display, HTML
# Graft our results matrix into pandas data frames
overlap_results_df = pd.DataFrame(data=overlap_results, index=["before registration", "after registration"],
columns=[name for name, _ in
OverlapMeasures.__members__.items()])
surface_distance_results_df = pd.DataFrame(data=surface_distance_results, index=["before registration", "after registration"],
columns=[name for name, _ in SurfaceDistanceMeasures.__members__.items()])
# Display the data as HTML tables and graphs
display(HTML(overlap_results_df.to_html(float_format=lambda x: '%.3f' % x)))
display(HTML(surface_distance_results_df.to_html(float_format=lambda x: '%.3f' % x)))
overlap_results_df.plot(kind='bar', rot=1).legend(bbox_to_anchor=(1.6,0.9))
surface_distance_results_df.plot(kind='bar', rot=1).legend(bbox_to_anchor=(1.6,0.9));
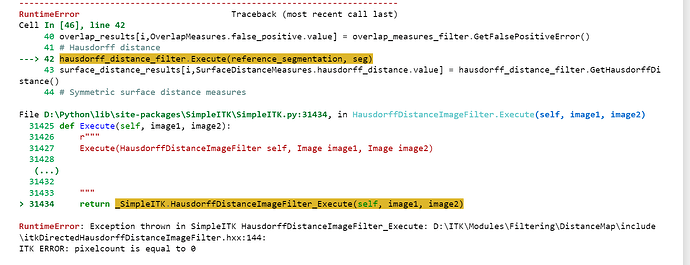
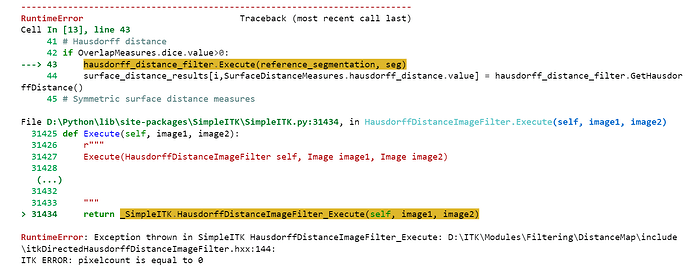
I am getting the following error.
What is the cause of this error? How can I solve this?