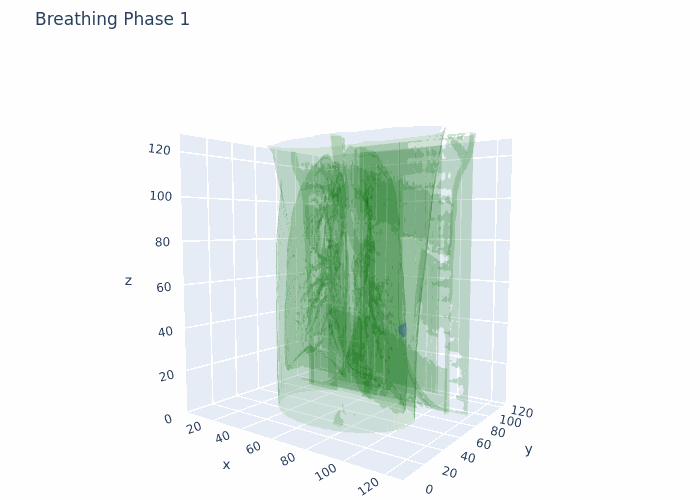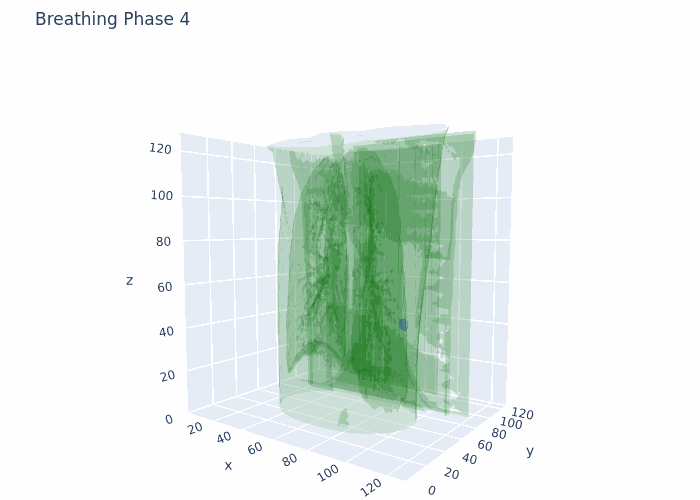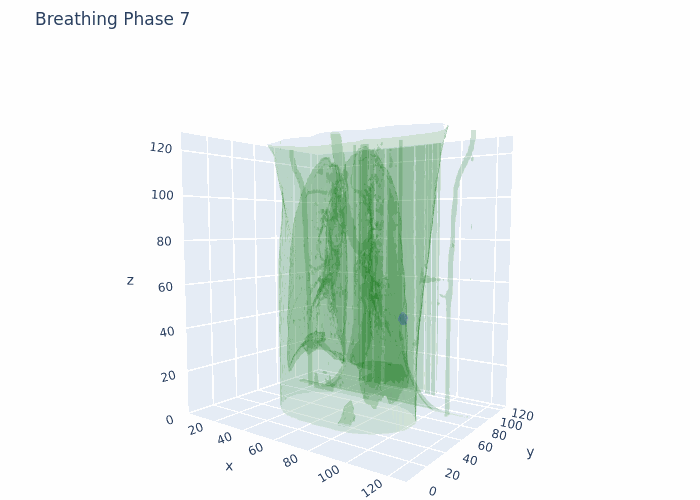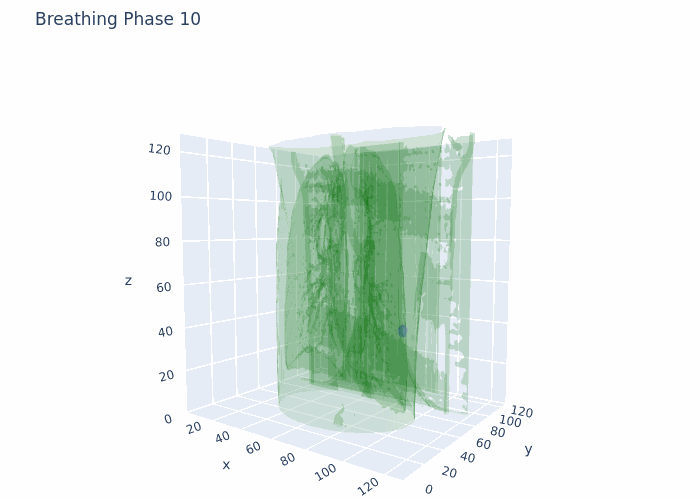
Hello,
I would like to ask for some guidance on how it would be possible to use deformation fields acquired from groupwise (or pairwise) registration to model (interpolate/simulate) the motion in-between frames.
Data that I have is 4D-CT 10 breathing phases, where for each breathing phase I have full chest CT (3D object) and full ROI (3D tumor mask). The shape is 10 (phases),128(height),128(width),128(depth) in 4D numpy array format. I also have amplitudes of chest during respiratory motion from a surface scanner signal at each time point from the full acquisition (continuous).
Now what I would like to achieve is to get from 10 breathing phases to, lets say for example, 90 to get a full breathing cycle. So I would like to interpolate 8 phases between original phase 1 and phase 2, then again interpolate 8 phases between original phase 2 and phase 3 and so on to get as smooth as possible motion. In addition, if possible, I would like to guide the interpolation based on the amplitudes that I have.
Basically simulating/modeling full respiratory motion during the 4D-CT acquisition.
Now focusing only on the ROIs-Tumor:
What I have tried out so far:
- I have applied groupwise registration to align all of the images to a common reference frame.
parameter_object = itk.ParameterObject.New()
groupwise_parameter_map = parameter_object.GetDefaultParameterMap('groupwise')
print("Original Metric:", groupwise_parameter_map['Metric'])
print("Original FinalGridSpacingInPhysicalUnits:", groupwise_parameter_map['FinalGridSpacingInPhysicalUnits'])
groupwise_parameter_map['Metric'] = ['AdvancedMattesMutualInformation']
groupwise_parameter_map['UseCyclicTransform'] = ['true']
groupwise_parameter_map['FinalGridSpacingInPhysicalUnits'] = ['64.0','64.0','64.0']
groupwise_parameter_map['Transform'] = ['AffineTransform']
groupwise_parameter_map['RegularizationType'] = ['Gaussian']
groupwise_parameter_map['RegularizationKernels'] = ['5']
parameter_object.AddParameterMap(groupwise_parameter_map)
elastix_object = itk.ElastixRegistrationMethod.New(image_itk_4D_tumor_extracted, image_itk_4D_tumor_extracted)
elastix_object.SetParameterObject(parameter_object)
elastix_object.SetLogToConsole(True)
elastix_object.UpdateLargestPossibleRegion()
result_image = elastix_object.GetOutput()
result_transform_parameters_tumor = elastix_object.GetTransformParameterObject()
numpy_array_tumor = itk.array_from_image(result_image)
-
I have validated the results visually and it provides somewhat satisfying results, the motion is quite okay and cyclical (as it should be).
-
Then I moved on to the interpolation, where I tried to first interpolate between two most distinct phases based on motion (distance difference):
fixed_image=itk.image_from_array(numpy_array_tumor[4])
moving_image=itk.image_from_array(numpy_array_tumor[0])
parameter_object = itk.ParameterObject.New()
parameter_map_rigid = parameter_object.GetDefaultParameterMap('affine')
parameter_map_rigid['Metric'] = ['PCAMetric2']
parameter_map_rigid['FinalGridSpacingInPhysicalUnits'] = ['4.0','4.0','4.0']
parameter_map_rigid['Transform'] = ['EulerTransform']
parameter_map_rigid['RegularizationType'] = ['Gaussian']
parameter_map_rigid['RegularizationKernels'] = ['2']
parameter_object.AddParameterMap(parameter_map_rigid)
result_image, result_transform_parameters = itk.elastix_registration_method(
fixed_image, moving_image,
parameter_object=parameter_object,
log_to_console=True)
deformation_field = itk.transformix_deformation_field(moving_image, result_transform_parameters)
deformation_field = itk.array_from_image(deformation_field)
image_itk_tumor = sitk.GetImageFromArray(numpy_array_tumor[4])
interpolated_images = []
num_steps = 8
index = 0
for step in tqdm(range(1, num_steps + 1)):
fraction = step / num_steps
scaled_deformation_field = deformation_field * fraction
scaled_deformation_field = sitk.GetImageFromArray(scaled_deformation_field.astype(np.float64),isVector=True)
interpolator = sitk.ResampleImageFilter()
interpolator.SetReferenceImage(image_itk_tumor)
interpolator.SetInterpolator(sitk.sitkBSpline)
interpolator.SetTransform(sitk.DisplacementFieldTransform(scaled_deformation_field))
interpolated_image = interpolator.Execute(image_itk_tumor)
interpolated_images.append(sitk.GetArrayFromImage(interpolated_image))
interpolated_images = np.stack(interpolated_images,axis=0)
- After validating the results visually, it seems that the motion is quite off:
a) It starts for some reason in between the phase 0 and phase 4 and not from phase 0 is it should.
b) It ends way above the phase 4 and not at the phase 4.
My questions would be:
- Am I going in the right direction based on my goal ?
- What could I do differently to make the motion more accurate ?
Sorry for the broad question, just looking for some guidance.
Attaching a GIF for reference with what I am working at the moment. This is original data, no registration and no interpolation done here.