Hi
I am trying to generate MIP for 3d brain mra images for aneurysm detection however I am struggling with which libraries to use and how to achieve this with python code
import os
import numpy as np
import SimpleITK as sitk
def make_mips(image_path, output_dir):
image = sitk.ReadImage(image_path)
image_size = image.GetSize()
basename = os.path.basename(image_path)
if not os.path.isdir(output_dir):
os.makedirs(output_dir)
for dim in range(3):
projection = sitk.MaximumProjection(image, dim)
if image_size[dim] % 2: # odd number
voxel = [0, 0, 0]
voxel[dim] = (image_size[dim] - 1) / 2
origin = image.TransformIndexToPhysicalPoint(voxel)
else: # even
voxel1 = np.array([0, 0, 0], int)
voxel2 = np.array([0, 0, 0], int)
voxel1[dim] = image_size[dim] / 2 - 1
voxel2[dim] = image_size[dim] / 2
point1 = np.array(image.TransformIndexToPhysicalPoint(voxel1.tolist()))
point2 = np.array(image.TransformIndexToPhysicalPoint(voxel2.tolist()))
origin = np.mean(np.vstack((point1, point2)), 0)
projection.SetOrigin(origin)
projection.SetDirection(image.GetDirection())
proj_basename = basename.replace('.nii.gz', '_mip_{}.nii.gz'.format(dim))
sitk.WriteImage(projection, os.path.join(output_dir, proj_basenam
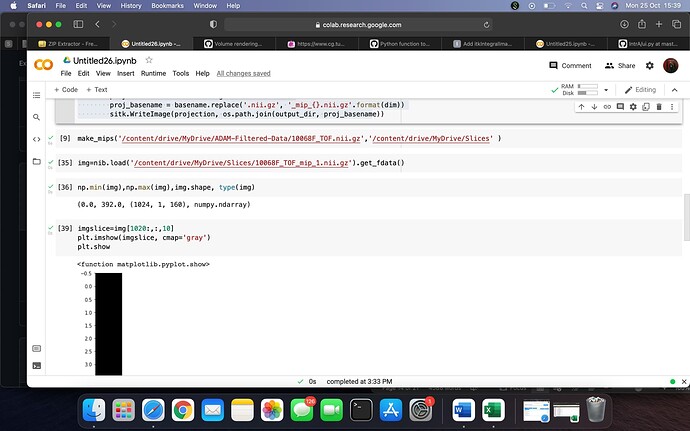
this code is generating blank images and not the MIP for brain vessels
Hello @Aleena_Suhail,
Welcome to SimpleITK!
There are a variety of projection filters (MaximumProjectionImageFilter is one of them). Note that the projection filters do not change the dimensionality of the result.
import SimpleITK as sitk
image = sitk.ReadImage('training_001_ct.mha')
# projection maintains original image dimensionality so output is [size_x,size_y,1]
# so we need to extract a 2D version
mip_axial = sitk.MaximumProjection(image, projectionDimension=2)[:,:,0]
sitk.Show(mip_axial, 'axial MIP')
I am getting Exception thrown in SimpleITK Show: /tmp/SimpleITK/Code/IO/src/sitkImageViewer.cxx:620:
sitk::ERROR: No ImageJ/Fiji application found.
this error
Is there a way to download the MIP as png format and view it without using imagj
Yes, just write it out (requires rescaling the intensities to [0,255]). Replace the sitk.Show with:
sitk.WriteImage(sitk.Cast(sitk.RescaleIntensity(mip_axial), sitk.sitkUInt8), 'axialMIP.png')
import SimpleITK as sitk
image = sitk.ReadImage('/content/drive/MyDrive/ADAM-Filtered-Data/10068F_TOF.nii.gz')
# projection maintains original image dimensionality so output is [size_x,size_y,1]
# so we need to extract a 2D version
mip_axial = sitk.MaximumProjection(image, projectionDimension=2)[:,:,0]
sitk.WriteImage(sitk.Cast(sitk.RescaleIntensity(mip_axial), sitk.sitkUInt8), 'axialMIP.png')
image = sitk.ReadImage("axialMIP.png”)
plt.imshow(image, cmap='gray')
plt.show
with this code I am getting the following error:
TypeError Traceback (most recent call last)
[<ipython-input-49-5973988eeef4>](https://xgeiymqwb-496ff2e9c6d22116-0-colab.googleusercontent.com/outputframe.html?vrz=colab-20211020-060059-RC00_404451640#) in <module>() ----> 1 plt.imshow(image, cmap='gray') 2 plt.show
5 frames
[/usr/local/lib/python3.7/dist-packages/matplotlib/image.py](https://xgeiymqwb-496ff2e9c6d22116-0-colab.googleusercontent.com/outputframe.html?vrz=colab-20211020-060059-RC00_404451640#) in set_data(self, A) 697 or self._A.ndim == 3 and self._A.shape[-1] in [3, 4]): 698 raise TypeError("Invalid shape {} for image data" --> 699 .format(self._A.shape)) 700 701 if self._A.ndim == 3:
TypeError: Invalid shape (1048576,) for image data
A SimpleITK image is not a numpy array.
You can get a numpy array via the sitk.GetArrayFromImage(image).
Thank you so much I have been able to generate MIP for one image I am trying to do the same for the whole dataset The code is executing without any error however it is not generating anything
This sounds like a Python programming problem, not ITK/SimpleITK. From the screenshot it appears that the WriteImage isn’t using a valid syntax. Possibly try:
sitk.WriteImage(sitk.Cast(sitk.RescaleIntensity(mip_axial),sitk.sitkUInt8), f'axialMIP{index}.png')
For future reference, please do not use screenshots for sharing code. Share a minimal working example of the code illustrating the specific problem.
dir = ‘content/drive/MyDrive/ADAM-Filtered-Data/’
output=’/content/drive/MyDrive/Slices’
for index,file_name in enumerate(sorted(os.listdir(’/content/drive/MyDrive/ADAM-Filtered-Data’))):
if file_name.endswith(‘TOF.nii.gz’):
image=sitk.ReadImage(’/content/drive/MyDrive/ADAM-Filtered-Data/’ + file_name)
mip_axial = sitk.MaximumProjection(image, projectionDimension=2)[:,:,0]
sitk.WriteImage(sitk.Cast(sitk.RescaleIntensity(mip_axial),sitk.sitkUInt8),output, f’axialMIP{index}.png’)
How can I modify this code so that the images are written in a particular file called ‘output’
Hello @Aleena_Suhail,
As the files are written in a loop, using the same file name will cause the images to overwrite each other, so in the end only the last image will be saved. If you want the file name to have a prefix of output instead of axialIMIP then just replace the text.
Note that this forum is for ITK/SimpleITK questions and not for general Python questions, which is what you appear to need. Please post Python questions in a more appropriate forum (stack overflow).