I am trying to register a segmented image on an MR image.
image shapes: (22, 20) (22, 20)
The MR image has intensity normalized from 0-1. The segmentation map has labels from 0-3(0 - background).
Both images need some rotational reorientation,
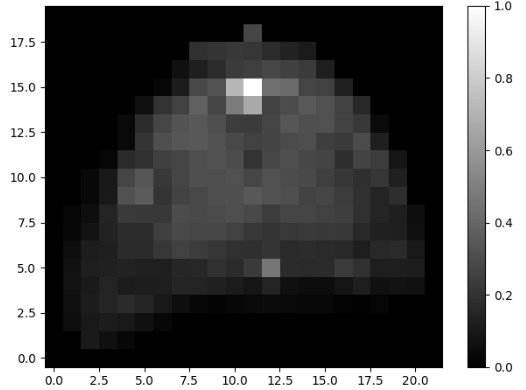
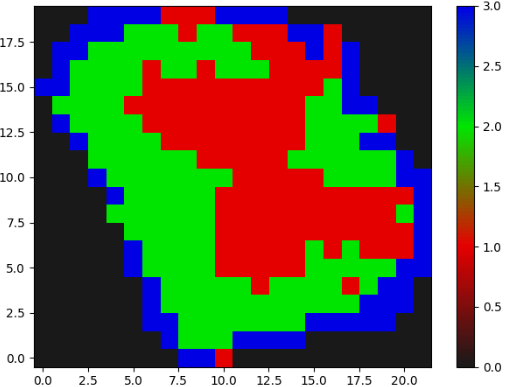
as can be seen above.
So I wanted to perform Euler transformation before delving into any non-rigid transformation.
I am using the following codes to do that:
mrItk = sitk.GetImageFromArray(mrImgNorm)
segItk = sitk.GetImageFromArray(secImg_resized)
fixed_image = sitk.Cast(mrItk, sitk.sitkFloat32)
moving_image = sitk.Cast(segItk, sitk.sitkFloat32)
print("image shapes: ", fixed_image.GetSize(), moving_image.GetSize())
all_orientations = np.linspace(-2 * np.pi, 2 * np.pi, 100)
# print(all_orientations)
# Evaluate the similarity metric using the rotation parameter space sampling, translation remains the same for all.
initial_transform = sitk.Euler2DTransform(sitk.CenteredTransformInitializer(fixed_image,
moving_image,
sitk.Euler2DTransform(),
sitk.CenteredTransformInitializerFilter.GEOMETRY))
# Registration framework setup.
registration_method = sitk.ImageRegistrationMethod()
registration_method.SetMetricAsMattesMutualInformation(numberOfHistogramBins=500)
registration_method.SetMetricSamplingStrategy(registration_method.RANDOM)
registration_method.SetMetricSamplingPercentage(0.001)
registration_method.SetInitialTransform(initial_transform, inPlace=False)
registration_method.SetOptimizerAsRegularStepGradientDescent(learningRate=2.0,
minStep=1e-4,
numberOfIterations=500,
gradientMagnitudeTolerance=1e-8)
registration_method.SetInterpolator(sitk.sitkNearestNeighbor)
registration_method.SetOptimizerScalesFromIndexShift()
# best_orientation = (0.0, 0.0)
best_similarity_value = registration_method.MetricEvaluate(fixed_image, moving_image)
similarity_value = []
# Iterate over all other rotation parameter settings.
for key, orientation in enumerate(all_orientations): # .items():
initial_transform.SetAngle(orientation)
registration_method.SetInitialTransform(initial_transform)
current_similarity_value = registration_method.MetricEvaluate(fixed_image, moving_image)
similarity_value.append(current_similarity_value)
# print("current similarity value: ", current_similarity_value)
if current_similarity_value <= best_similarity_value:
best_similarity_value = current_similarity_value
best_orientation = orientation
# else:
# best_orientation = orientation
print('best orientation is: ' + str(best_orientation))
print(current_similarity_value)
plt.plot(all_orientations, similarity_value, 'b')
plt.plot(best_orientation, best_similarity_value, 'rv')
plt.show()
initial_transform.SetAngle(best_orientation)
registration_method.SetInitialTransform(initial_transform, inPlace=False)
eulerTx = registration_method.Execute(fixed_image, moving_image)
resampler = sitk.ResampleImageFilter()
resampler.SetReferenceImage(fixed_image)
resampler.SetInterpolator(sitk.sitkNearestNeighbor)
resampler.SetDefaultPixelValue(0)
resampler.SetTransform(eulerTx)
out = resampler.Execute(moving_image)
displayImage(sitk.GetArrayFromImage(out), Title_='registered image(Euler2D)')
displayImage(sitk.GetArrayFromImage(moving_image), Title_='moving image')
displayImage(sitk.GetArrayFromImage(fixed_image), Title_='fixed image')
checkerImg = compare_images(sitk.GetArrayFromImage(out),
sitk.GetArrayFromImage(fixed_image),
method='diff')
displayImage(checkerImg, Title_='mask-label overlapped')
del registration_method, initial_transform, eulerTx
moving_image = sitk.Cast(out, sitk.sitkFloat32)
but I am getting this error at this line here:
ITK ERROR: MattesMutualInformationImageToImageMetricv4(00000207A2415B70): VirtualSampledPointSet must have 1 or more points.
eulerTx = registration_method.Execute(fixed_image, moving_image)
From a previous discussion here, I thought the image size must be equal for both the images, that is why I mentioned them here.
To some extent plotting the metric makes sense:
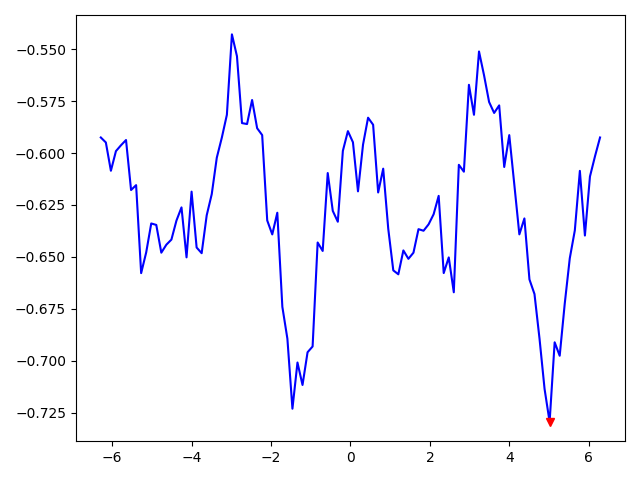
because at best_orientation (the red marker) the moving image seems to be somewhat matching with MR:
rotated_segmentation = ndimage.rotate(secImg_resized, np.rad2deg(best_orientation))
displayImage(rotated_segmentation, Title_='rotated sec')
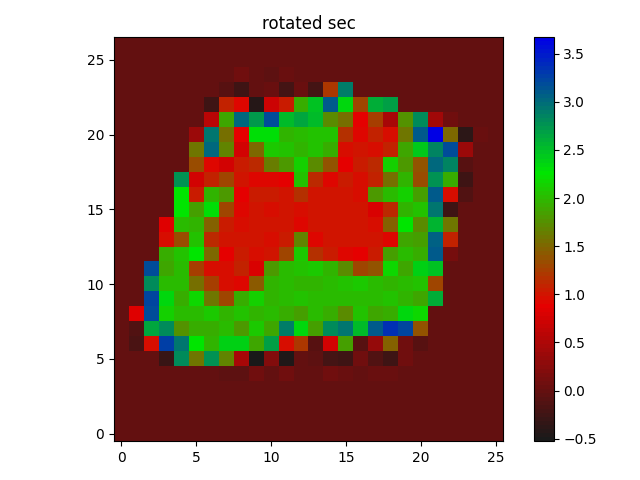
Obviously, the labels are changed, but this is not registered anyway, just rotating the image using ndimage simply.
what shall I do from here?