P.P.S. It looks like only inp0.npy is causing the “H&E getting mixed up” error. Each of the other inputs “seems to change all pixels to shades of pink” – I can take a look at that too. Thanks, Lee.
Hi Lee,
The attached slightly modified demo ITK color normalization code, ‘pp.py’, is changed to use the attached image file inputs and demonstrates my current problem. It was executed on my MacOS UNIX-like system as:
$ python3 pp.py 2>&1 | tee log_demo_mod
The python script ‘pp.py’, output ‘log_demo_mod’, reference file 'ref0.npy’ and input file 'inp0_(2,1).npy’ are attached.
The error message has the suggestions:
…
Possible solutions:
- If you are an application user:
** Convert your input image into a supported format (see below).
** Contact developer to report the issue. - If you are an application developer, force input images to be
loaded in a supported pixel type.
…
But the code prints out both the input and reference image types as '<class 'itk.itkImagePython.itkImageUC3’> which would seem to be a supported type.
So I’m at a loss as to what to do except ‘Contact developer to report the issue’.
The training and test data is all in numpy .npy format converted from the original .jpeg. There are ~30k 208x208 RGB “patches” taken from 1024x1024 “tiles” at 10x magnification†, themselves extracted from H&E whole slide images (WSIs) 10-50k pixels on a side.
Using the test data, the trained inference measures are really quite good. The physicians involved would like validation for their use to be performed on older data (and thus faded H&E). The RGB tiles are at least 102410243 = 3.15 Mb and so exceed the 3 Mb ITK email limit of 3 Mb. So I’m using smaller ‘patches’ in what I’m sending you with the input data the same size as the reference data, 208x208x3 pixels.
† https://wiki.cancerimagingarchive.net/x/xwElAw
Jon R. Sauer
jon.sauer@gmail.com
Acton, MA, USA 01720
+1 303.579.3009
pp.py (2.98 KB)
log_demo_mod (3.17 KB)
ref0.npy (127 KB)
inp0_(2,1).npy (127 KB)
We are no longer getting Hematoxylin and Eosin mixed up; maybe that is a good sign!
Because we are reading two-dimensional RGB images rather than three-dimensional monochromatic images, we need to use is_vector=True as in
input_image = itk.image_from_array(input_image, is_vector=True)
reference_image = itk.image_from_array(reference_image, is_vector=True)
With that fix, how does the code do on your platform? I am not getting any errors here.
Hi Lee,
Thanks. No errors also here. Now I can apply your color normalization to quite faded H&E Boston Children’s Hospital data and see if the model trained on the public-domain UTexas data gives reasonable results.
Again, many thanks for your very prompt and very useful reply‼️
Cheers 
 ,
,
Jon
Jon R. Sauer
jon.sauer@gmail.com
Acton, MA, USA 01720
+1 303.579.3009
I am glad that I was able to help. If you have further problems… you know how to reach me. Peace --Lee.
Hi Lee,
Great that color normalization works sometimes. But on just my 2nd test input ("inp0_(0,3).npy”, attached), it fails 
Error ends with
“ITK ERROR: The image to be normalized could not be processed; does it have white, blue, and pink pixels?”
To my uneducated eye, I didn’t see the two test inputs as being really different. Perhaps I need to filter the input files in some way?
The change to the test script is seen below:
input_image_filename = ‘inp0_(2,1).npy’ # source tile with patch coords
goes to
#input_image_filename = ‘inp0_(2,1).npy’ # source tile with patch coords, works post Lee fix 220217
input_image_filename = ‘inp0_(0,3).npy’ # source tile with patch coords
Jon R. Sauer
jon.sauer@gmail.com
Acton, MA, USA 01720
+1 303.579.3009
inp0_(0,3).npy (127 KB)
Hi Lee,
The attached python3 script “pp.py" uses the remaining attached files (the last one for reference, six for input (so under 1Mb)) to the color normalization code. Sometimes it succeeds (especially on "inp0_(2,1).npy”, but for the others it mostly fails with errors:
ITK ERROR: The image to be normalized could not be processed; does it have white, blue, and pink pixels?
and
ITK ERROR: Hematoxylin and Eosin are getting mixed up; failed .
Running the script multiple times shows the same input, when it fails, seemingly randomly switching between these two failure types and even succeeding sometimes.
I think this is because the H&E staining is faded. If you also think this, is there some preprocessing on faded H&E inputs that you can recommend? Or is there a switch I can use to increase the success rate?
I run the script (not using jupyter notebooks) as
$ python3 pp.py 2>log_errs_pp
and exiting each matplotlib plot to continue.
I think this is because the H&E staining is faded. If you also think this, is there some preprocessing on faded H&E inputs that you can recommend?
The “tiles” are 1024x1024 pixel subsections of much larger whole slide images converted to numpy arrays filtered to eliminate mostly blank or badly mis-stained areas. The “patches” are smaller (208x208 pixel) tile subsections large enough to allow classification (in most cases) but small enough to allow rapid filtering/preprocessing and training. These sort of splits are in common use in the community.
Let me know if you at least get this message.
Thanks.
Cheers,
Jon
Jon R. Sauer
jon.sauer@gmail.com
Acton, MA, USA 01720
+1 303.579.3009
pp.py (5.84 KB)
inp0_(0,3).npy (127 KB)
inp0_(1,1).npy (127 KB)
inp0_(1,3).npy (127 KB)
inp0_(2,1).npy (127 KB)
inp0_(2,3).npy (127 KB)
inp0_(3,0).npy (127 KB)
ref0.npy (127 KB)
Sometimes with this reference
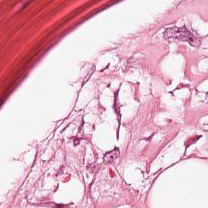
and this input
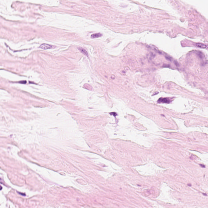
the no-error result is

which is clearly wrong.
Hopefully you have a path forward to attack this problem.
Cheers,
Jon
Jon R. Sauer
jon.sauer@gmail.com
Acton, MA, USA 01720
+1 303.579.3009
Short update: I have been working on this occasionally and hence slowly. I have uncovered that a sanity check that should have failed earlier in the process was not properly written. I fixed that, so I am now failing earlier with your test case from a few days ago. I haven’t yet figured out what is causing that failure.
Hi Lee,
Thanks for update.
No rush, take your time. I was an experimental particle physicist for decades. Building hardware always seemed more satisfying than debugging software. Errors easier to find and when fixed, testing usually straightforward and the fix was more likely to be robust in many cases.
But things can go literally explosively wrong. It was my experiment on a hydrogen bubble chamber in 1965 that blew up killing a young technician and putting Harvard’s Cambridge Electron Accelerator out of commission for over a year. I was just entering the building at 3:30am.
I’m testing the package ’staintools’ from Peter Byfield now. In it’s simplest form, straight from his example, it seems to work better for the pink eosin stains, not so sure yet about the purplish-blue hematoxylin stains.
Cheers,
Jon
Jon R. Sauer
jon.sauer@gmail.com
Acton, MA, USA 01720
+1 303.579.3009
This fix is being released and will be available as pip install -U 'spcn>=0.1.7' shortly. It should have been released overnight but there was some sort of unrelated hiccough in the process, so I have just restarted it.
I just corrected the previous post to indicate itk-spcn rather than the typo spcn.
It is released. Please try
pip install -U 'itk-spcn>=0.1.7'
Hi Lee —
Many, many thanks. Works MUCH better now.
Cheers,
Jon
Jon R. Sauer
jon.sauer@gmail.com
Acton, MA, USA 01720
+1 303.579.3009
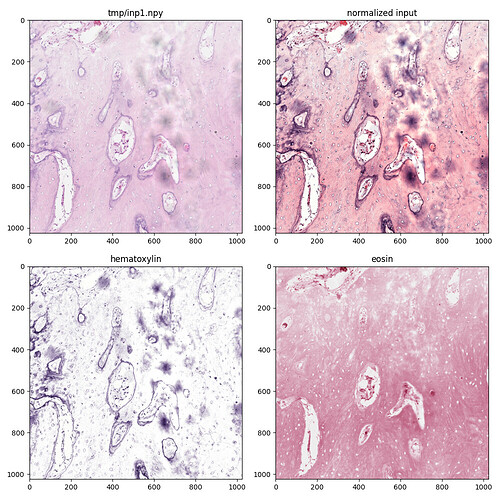
Hi Lee,
In the meantime, I used a Python version of the original Macenko MATLAB code to see how it worked. It hard-codes the reference stain parameters (but I couldn’t find where they came from) and so is less appealing and less flexible than your package. But it allows me to see what is used in the normalized output for each input stain, hematoxylin and eosin, as attached.
Is there a way to get at these separated images in your package?
Cheers,
Jon
Jon R. Sauer
jon.sauer@gmail.com
Acton, MA, USA 01720
+1 303.579.3009
I have submitted ITKColorNormalization Issue #32 to request this functionality.
Hi Lee,
Thanks. The reason this would be useful to me is to better convince myself, and others (in this case physicians), that the code is working exactly as advertised. The extra output would only be used in testing or demonstration scenarios so, just as you say, only when requested by the user.
Cheers,
Jon
Jon R. Sauer
jon.sauer@gmail.com
Acton, MA, USA 01720
+1 303.579.3009