Hi Lee,
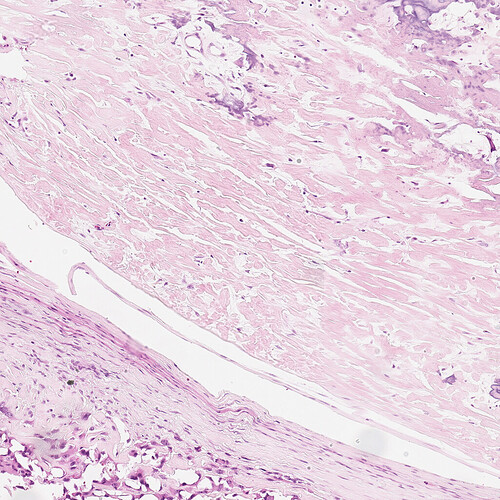
The image included below is a typical “tile” (small section of a much larger whole slide image of Boston Children’s Hospital (BCH) H&E stained osteosarcoma resected bone) of the data I’d like to apply structure-preserving color normalization to. These tiles are nearly all under-stained compared to the reference image used in the example code or the well-curated public domain images I trained a CNN on.
What I was hoping to do is use the ITK Color Normalization facility to make the BCH data more like the well-stained training data or the example reference image which are very similar.
The code below is taken from
https://github.com/InsightSoftwareConsortium/ITKColorNormalization/blob/master/examples/ITKColorNormalization.ipynb
with a few slight modifications to optionally run on my iTerm command line environment instead of the original Jupyter notebook (“JRS=True”) or use a BCH tile (“BCH=True”).
I hope it won’t be too hard for you to reproduce my problems.
The diffs from the example code are in “diffs.txt" below. The BCH image is in “Test_1_0810.png”. The actual code used is in “xx.py”. The errors are in “errs.txt”.
diffs.txt (1.99 KB)
xx.py (3.78 KB)
errs.txt (904 Bytes)