Hi,
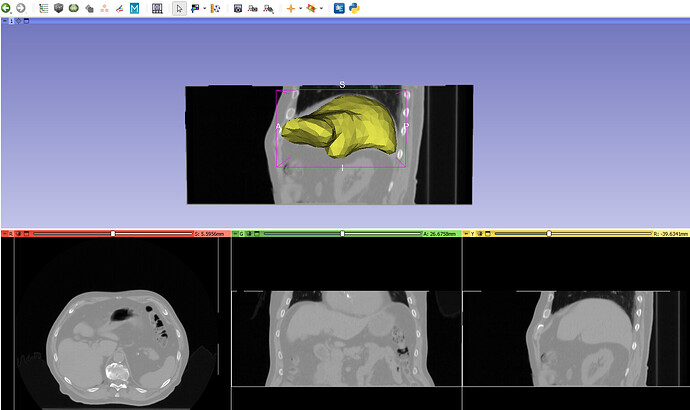
I have generated deformed 3D-CT versions and corresponding deformation vector field (DVF) volumes of a given nifti image volume (reference image 3D-CT volume) using another tool. I then extracted the liver mesh from the binary mask image volume obtained by segmenting the liver region from the reference image volume. They are perfectly align one another when I visualized it through using the 3D slicer.
Next, I apply corresponding DVF by first extracting displacement field and then apply the transform matrix to each and every point in the reference mesh to deform so that they also be able to align with the corresponding deformed CT volumes (you can see the code below).
(This means that given a physical point and displacement vector field, I apply the transformation defined by the displacement field to that point.)
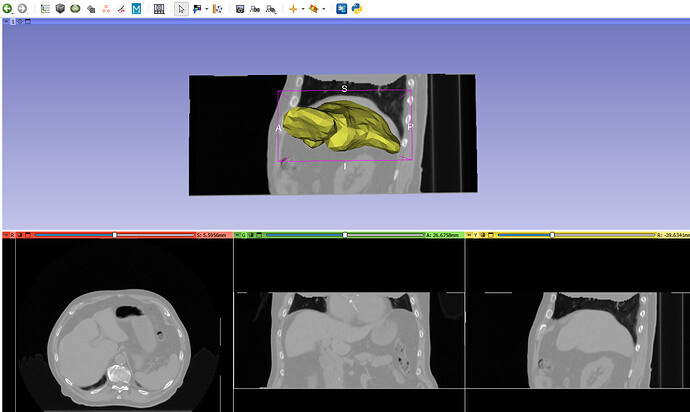
However, I’ve encountered an issue where, in some deformed instances, the corresponding deformed mesh does not align as expected. I suspect that this discrepancy could be attributed to linear interpolation during the TransformPoint process using Simple ITK. I’m keen to understand if there’s a solution for this issue or if there’s something I might have done wrong in the process ?
For further context, I’ve attached sample data which includes two deformed 3D CTs and their corresponding DVFs. The first case demonstrates correct deformation, while the second does not. Additionally, I’ve included a visualization in 3D Slicer for reference.
I would greatly appreciate any assistance to address this concern and resolve this alignment issue. Your insights would be invaluable in helping me to resolve this issue.
def generateDeformedMeshesFromITKDVFs(data_dir, reference_image_physical_points, reference_mesh):
folder_list = os.listdir(data_dir)
for i, folder_name in enumerate(folder_list):
main_folder_path = os.path.join(data_dir, folder_name)
dvf_folder_path = os.path.join(main_folder_path, 'ITK_DVFs')
# create a folder to save deformed meshes for each case
deformed_mesh_path = os.path.join(main_folder_path, 'deformedMeshes')
if(not os.path.exists(deformed_mesh_path)):
os.mkdir(deformed_mesh_path)
dvf_vol_names = [x for x in os.listdir(dvf_folder_path) if x.endswith(".nii.gz")]
for j, relative_dvf_path in enumerate(dvf_vol_names):
dvf_img_path = os.path.join(dvf_folder_path, relative_dvf_path)
# read dvf image volume which is in nifti format with dim 5
dvf_image = sitk.ReadImage(dvf_img_path)
corrected_dvf_image = sitk.Compose(
[
sitk.VectorIndexSelectionCast(dvf_image, 0, sitk.sitkFloat64),
sitk.VectorIndexSelectionCast(dvf_image, 1, sitk.sitkFloat64),
sitk.VectorIndexSelectionCast(dvf_image, 2, sitk.sitkFloat64),
]
)
#create the SimpleITK representation of the DVF transformation, which differs from the dvf_image
# generate displacement vector field
sitk_dvf = sitk.DisplacementFieldTransform(reference_image_physical_points - corrected_dvf_image)
reference_mesh_duplicate = copy.deepcopy(reference_mesh)
transformed_physical_points = []
# transform each point in the mesh using a displacement field
for index in range(0, reference_mesh_duplicate.points.shape[0]):
transformed_physical_points.append(sitk_dvf.TransformPoint(reference_mesh_duplicate.points[index]))
# convert the list of tuples into 2D numpy array
list_tuples_to_numpy_2d_array = np.array([*transformed_physical_points])
# assign trasnformed points as the new mesh points to teh copy of a reference mesh
reference_mesh_duplicate.points = list_tuples_to_numpy_2d_array
dvf_img_name_split = relative_dvf_path.split('DVF', 1)
new_mesh_name_with_ext = dvf_img_name_split[0] + 'simDyn' + dvf_img_name_split[1].rsplit('.', 2)[0] + '.vtk'
new_mesh_save_path = os.path.join(deformed_mesh_path, new_mesh_name_with_ext)
# write new mesh to the disk using meshio
meshio.write(new_mesh_save_path, reference_mesh_duplicate, binary=False)
reference_image = sitk.ReadImage('./SIMPLE_ITK_SAMPLE_DATA_MESH_INTERPOALTION/reference_image/MCR.nii.gz')
reference_image_physical_points = sitk.PhysicalPointSource(outputPixelType = sitk.sitkVectorFloat64,
size = reference_image.GetSize(),
origin = reference_image.GetOrigin(),
spacing = reference_image.GetSpacing(),
direction = reference_image.GetDirection())
reference_mesh = meshio.read('./SIMPLE_ITK_SAMPLE_DATA_MESH_INTERPOALTION/reference_mesh/MCR.vtk')
datadir = './SIMPLE_ITK_SAMPLE_DATA_MESH_INTERPOALTION/deformed_cts_dvfs'