I made an experiment about the itk.imread function, and found something interesting.
I putted .dcm files with the same series ID in a directory. And then convert the dicom data into nii.gz format. Here come the difference:
Code-1: random shuffled files
import os
import itk
import random
dir_path = "./20210718_010_NECK_T1W_mDIXON_TRA+C/"
results_path = "./results/"
filenames = os.listdir(dir_path)
random.shuffle(filenames)
filenames = [os.path.join(dir_path, f) for f in filenames]
image = itk.imread(filenames)
itk.imwrite(image, results_path + "image_shuffle.nii.gz")
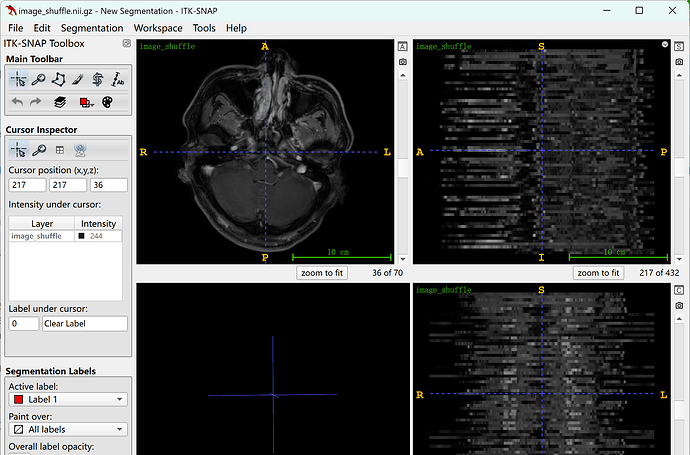
Here is the result visualized in ITK-SNAP:
It obviously showed a chaotic scene.
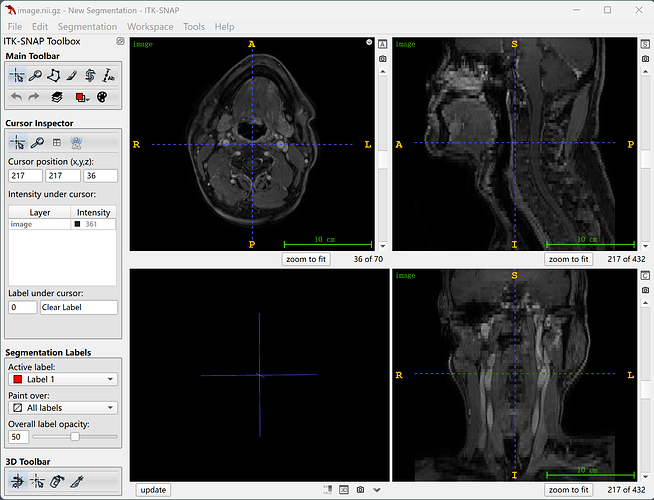
But if I try to convert it by passing a dir parameter, it worked well.
Code-2:
import itk
dir_path = "/mnt/pc/Chenq_team/HuangData/MR/935294/20210718_010_NECK_T1W_mDIXON_TRA+C/"
results_path = "./results/"
image = itk.imread(dir_path)
itk.imwrite(image, results_path + "image.nii.gz")
In fact, I didn’t mean to bring in random to say the there is a bug in ITK.
Instead, I encountered a similar scenario while processing the dicom data. In order to describe my problem, I gave the above two simple examples.
So my question is: Why does itk.imread affected by the order of passing file_list parameter?