Hello,
The problem that I am trying to solve with SimpleITK:
1/ I have a temporal sequence of volumes from the same patient
2/ I have one segmentation mask of the organ under observation.
3/ I want to get segmentations based on registration and transforms from the one mask for all the temporal sequences:
4/ Apply that for meshes
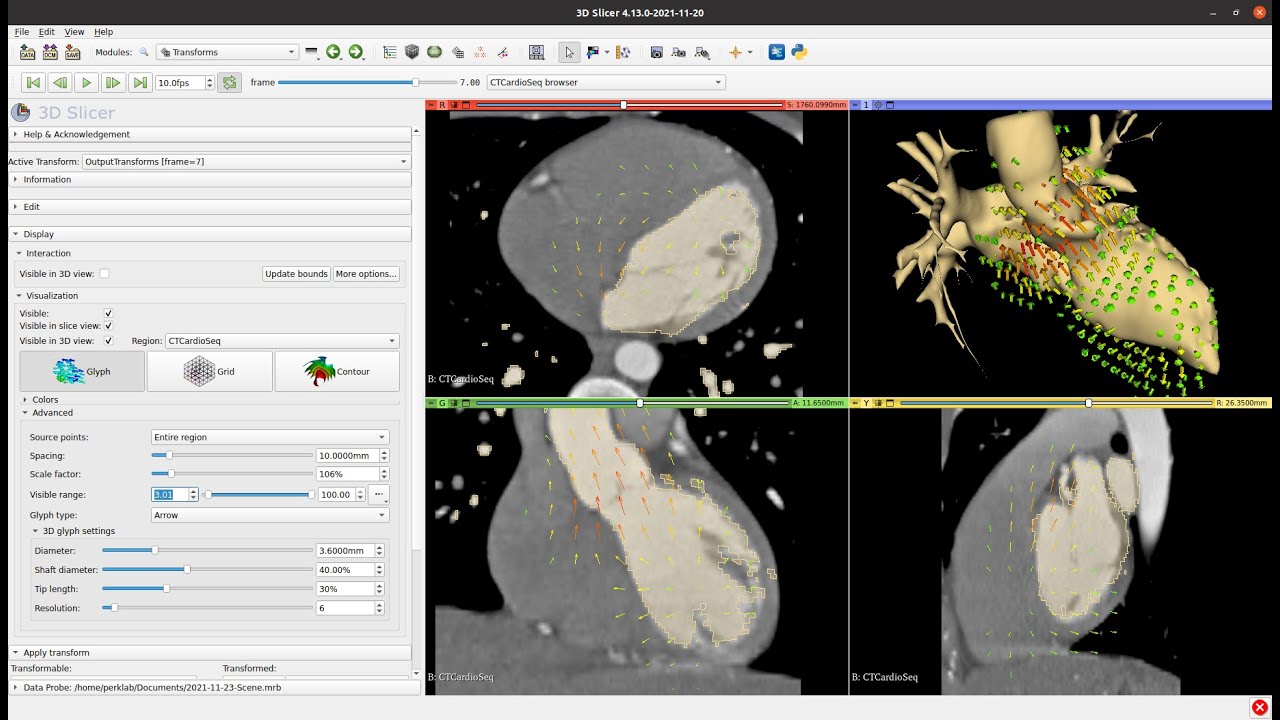
I first obtain the deformation fields and then warp the mask with the displacement field for all the volumes in the temporal sequence. Therefore I gain the segmentations for all volumes.
Vref = sitk.ReadImage(original_mask)
dvf_sitk = sitk.ReadImage(deformation_field_path)
resampler = sitk.ResampleImageFilter()
resampler.SetReferenceImage(Vref)
dis_tx = sitk.DisplacementFieldTransform(sitk.Cast(dvf_sitk,sitk.sitkVectorFloat64))
resampler.SetTransform(dis_tx)
out = resampler.Execute(Vref)
However, I want to work with meshes. For the first segmented volume I applied Delaunay surface triangulation to obtain the mesh. Then, I’d like to change the mesh based on the deformation field (therefore not changing the adjacency matrix and cardinality of the set of nodes).
I thought about using the displacementfield, however they yield float values.
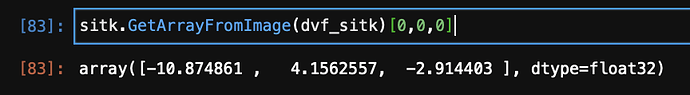
np.unique(sitk.GetArrayFromImage(dvf_sitk))
array([-33.68408 , -33.51517 , -33.3977 , ..., 19.781471, 19.782059, 19.919436], dtype=float32)
The documentation for ResampleImageFilter / WarpImageFilter states:
Note that the choice of interpolator function can be important. This function is set via SetInterpolator() . The default is LinearInterpolateImageFunction <InputImageType, TInterpolatorPrecisionType>, which is reasonable for ordinary medical images. However, some synthetic images have pixels drawn from a finite prescribed set.
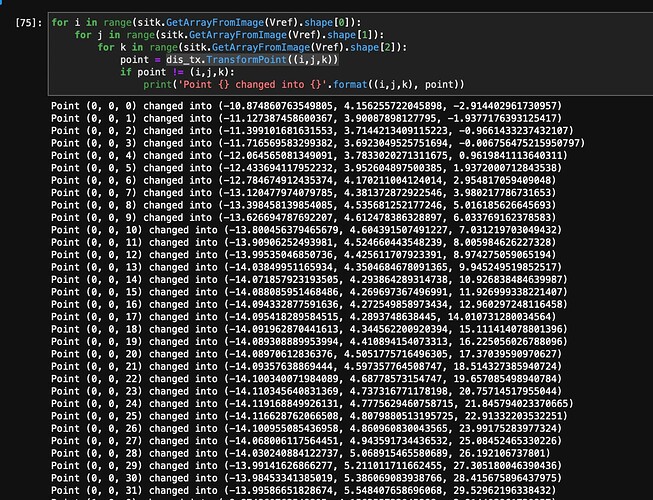
So, the warping after using the values from displacementfield interpolates the float numbers into integers. Since I want to work with mesh, how can I get the mappings how each point changed?
For instance, voxel [3,5,7] after resampling can be found at [3,5,8] etc.
Thanks!