Hi,
Here are the actions I took to resolve the orientation problem.
When I use the SimpleITK tool-kit to load the reference CT (i.e. RAS oriented MCR.nii.gz), the original orientation, i.e. in RAS, changes to LPS orientation. For example, when I print the physical boundaries, I get the numbers shown below.
physical point at voxel [0, 0, 0]: (444.0, 380.0, -320.0)
physical point at voxel [202, 126, 160]: (40.0, 128.0, 0.0)
Image Origin: (444.0, 380.0, -320.0)
Voxel Spacing: (2.0, 2.0, 2.0)
Image Direction: (-1.0, 0.0, 0.0, 0.0, -1.0, 0.0, 0.0, 0.0, 1.0)
Image Size: (203, 127, 161)
As a result, I modified the origin and direction of the reference CT to RAS because ITK loads an LPS-oriented image by default, as shown in the steps below. Once the previously mentioned information has been modified, save it using the code below.
reference_image = sitk.ReadImage('./MCR.nii.gz')
ref_image_out = sitk.GetImageFromArray(sitk.GetArrayFromImage(reference_image))
ref_image_out.SetOrigin((-reference_image.GetOrigin()[0], -reference_image.GetOrigin()[1], reference_image.GetOrigin()[2]))
ref_image_out.SetSpacing(reference_image.GetSpacing())
ref_image_out.SetDirection((1.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 1.0))
sitk.WriteImage(ref_image_out, './ITK_RAS/MCR.nii.gz')
physical point at voxel [0, 0, 0]: (-444.0, -380.0, -320.0)
physical point at voxel [202, 126, 160]: (-40.0, -128.0, 0.0)
Image Origin: (-444.0, -380.0, -320.0)
Voxel Spacing: (2.0, 2.0, 2.0)
Image Direction: (1.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 1.0)
Image Size: (203, 127, 161)
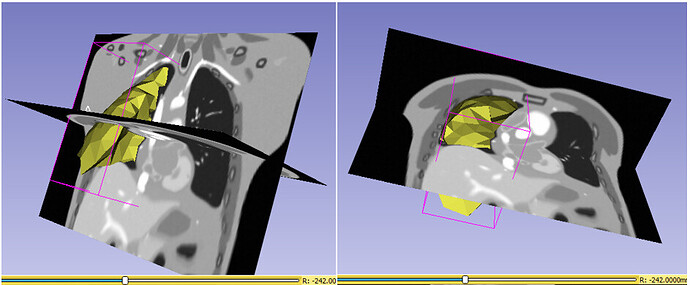
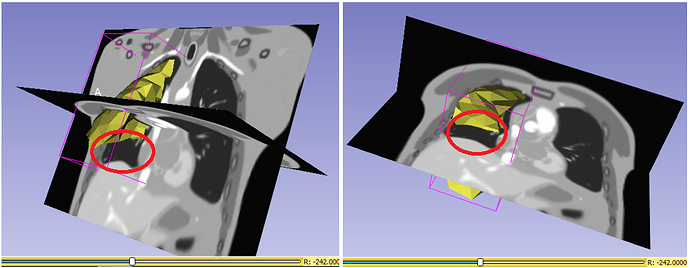
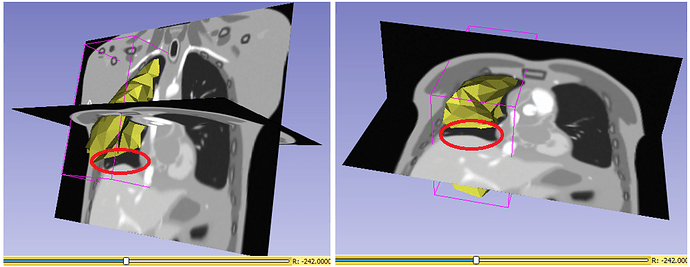
All of the mesh points are now within the domain of the displacement field transformation as a result of this change. I can guarantee you of this since the mesh points are now properly aligned with the reference CT volume. Furthermore, I applied the same changes to the deoformed CT volumes and DVF image volumes.
However, as I stated in the previous thread, the output mesh points after applying the displacement fields are incorrectly distorted, presumably because they are not aligned with slices in the associated deformed image volume, as I indicated above. Should I apply inverse of that transform ? or am I mssing something ?
If possible, could please help me to resolve this issue.