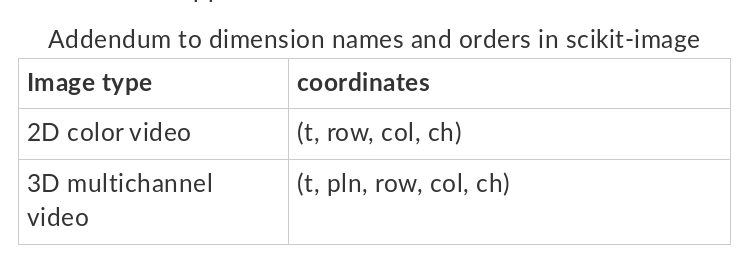
It is reasonable to apply the conventions of scikit-image for indexing, space, channel, and time order. These are compatible with how data is natively organized in ITK and NumPy array interfaces to itk.Image. That is, [k,j,i] indexing,
We can add documentation to a repository and use pull requests and line-based comments for additional discussion.