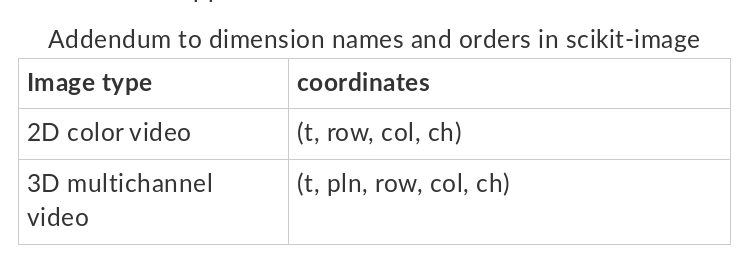
Here are few examples! 
They,
- Generate correct scales and translations, aka as spacing and origin in ITK lingo, which are included in v0.4 of the OME-Zarr specification. This includes correct values for the multiple scales.
- Generate the multiscale representation for the first and second library
- Can be used in memory in Python or to convert files from ITK and non-ITK-based IO libraries, tifffile, imageio, or pyimagej, preserving or adding spatial metadata as needed.
 Features
Features
- Minimal dependencies
- Work with arbitrary Zarr store types
- Lazy, parallel, and web ready – no local filesystem required
- Process extremely large datasets
- Multiple downscaling methods
- Supports Python>=3.8
- Implements version 0.4 of the
OME-Zarr NGFF specification
Installation
To install the command line interface (CLI):
pip install 'ngff-zarr[cli]'
CLI example:
ngff-zarr -i ./MR-head.nrrd -o ./MR-head.ome.zarr
Bidirectional type conversion that preserves spatial metadata is available with
itk_image_to_ngff_image and ngff_image_to_itk_image.
Once represented as an NgffImage, a multiscale representation can be generated
with to_multiscales. And an OME-Zarr can be generated from the multiscales
with to_ngff_zarr.
ITK Python
An example with
ITK Python:
>>> import itk
>>> import ngff_zarr as nz
>>>
>>> itk_image = itk.imread('cthead1.png')
>>>
>>> ngff_image = nz.itk_image_to_ngff_image(itk_image)
>>>
>>> # Back again
>>> itk_image = nz.ngff_image_to_itk_image(ngff_image)
ITK-Wasm Python
An example with ITK-Wasm. ITK-Wasm’s Image is a simple
Python dataclass like NgffImage.
>>> from itkwasm_image_io import imread
>>> import ngff_zarr as nz
>>>
>>> itk_wasm_image = imread('cthead1.png')
>>>
>>> ngff_image = nz.itk_image_to_ngff_image(itk_wasm_image)
>>>
>>> # Back again
>>> itk_wasm_image = nz.ngff_image_to_itk_image(ngff_image, wasm=True)
Python Array API
NGFF-Zarr supports conversion of any NumPy array-like object that follows the
Python Array API Standard into
OME-Zarr. This includes such objects an NumPy ndarray’s, Dask Arrays, PyTorch
Tensors, CuPy arrays, Zarr array, etc.
Array to NGFF Image
Convert the array to an NgffImage, which is a standard
Python dataclass that
represents an OME-Zarr image for a single scale.
When creating the image from the array, you can specify
- names of the
dims from {‘t’, ‘z’, ‘y’, ‘x’, ‘c’}
- the
scale, the pixel spacing for the spatial dims
- the
translation, the origin or offset of the center of the first pixel
- a
name for the image
- and
axes_units with
UDUNITS-2 identifiers
>>> # Load an image as a NumPy array
>>> from imageio.v3 import imread
>>> data = imread('cthead1.png')
>>> print(type(data))
<class 'numpy.ndarray'>
Specify optional additional metadata with to_ngff_zarr.
>>> import ngff_zarr as nz
>>> image = nz.to_ngff_image(data,
dims=['y', 'x'],
scale={'y': 1.0, 'x': 1.0},
translation={'y': 0.0, 'x': 0.0})
>>> print(image)
NgffImage(
data=dask.array<array, shape=(256, 256),
dtype=uint8,
chunksize=(256, 256), chunktype=numpy.ndarray>,
dims=['y', 'x'],
scale={'y': 1.0, 'x': 1.0},
translation={'y': 0.0, 'x': 0.0},
name='image',
axes_units=None,
computed_callbacks=[]
)
The image data is nested in a lazy dask.array and chucked.
If dims, scale, or translation are not specified, NumPy-compatible
defaults are used.
Generate multiscales
OME-Zarr represents images in a chunked, multiscale data structure. Use
to_multiscales to build a task graph that will produce a chunked, multiscale
image pyramid. to_multiscales has optional scale_factors and chunks
parameters. An antialiasing method can also be prescribed.
>>> multiscales = nz.to_multiscales(image,
scale_factors=[2,4],
chunks=64)
>>> print(multiscales)
Multiscales(
images=[
NgffImage(
data=dask.array<rechunk-merge, shape=(256, 256), dtype=uint8,chunksize=(64, 64), chunktype=numpy.ndarray>,
dims=['y', 'x'],
scale={'y': 1.0, 'x': 1.0},
translation={'y': 0.0, 'x': 0.0},
name='image',
axes_units=None,
computed_callbacks=[]
),
NgffImage(
data=dask.array<rechunk-merge, shape=(128, 128), dtype=uint8,
chunksize=(64, 64), chunktype=numpy.ndarray>,
dims=['y', 'x'],
scale={'x': 2.0, 'y': 2.0},
translation={'x': 0.5, 'y': 0.5},
name='image',
axes_units=None,
computed_callbacks=[]
),
NgffImage(
data=dask.array<rechunk-merge, shape=(64, 64), dtype=uint8,
chunksize=(64, 64), chunktype=numpy.ndarray>,
dims=['y', 'x'],
scale={'x': 4.0, 'y': 4.0},
translation={'x': 1.5, 'y': 1.5},
name='image',
axes_units=None,
computed_callbacks=[]
)
],
metadata=Metadata(
axes=[
Axis(name='y', type='space', unit=None),
Axis(name='x', type='space', unit=None)
],
datasets=[
Dataset(
path='scale0/image',
coordinateTransformations=[
Scale(scale=[1.0, 1.0], type='scale'),
Translation(
translation=[0.0, 0.0],
type='translation'
)
]
),
Dataset(
path='scale1/image',
coordinateTransformations=[
Scale(scale=[2.0, 2.0], type='scale'),
Translation(
translation=[0.5, 0.5],
type='translation'
)
]
),
Dataset(
path='scale2/image',
coordinateTransformations=[
Scale(scale=[4.0, 4.0], type='scale'),
Translation(
translation=[1.5, 1.5],
type='translation'
)
]
)
],
coordinateTransformations=None,
name='image',
version='0.4'
),
scale_factors=[2, 4],
method=<Methods.ITKWASM_GAUSSIAN: 'itkwasm_gaussian'>,
chunks={'y': 64, 'x': 64}
)
The Multiscales dataclass stores all the images and their metadata for each scale according the OME-Zarr data model. Note that the correct scale and translation for each scale are automatically computed.
Write to Zarr
To write the multiscales to Zarr, use to_ngff_zarr.
nz.to_ngff_zarr('cthead1.ome.zarr', multiscales)
Use the .ome.zarr extension for local directory stores by convention.
Any other Zarr store type can also be used.
The multiscales will be computed and written out-of-core, limiting memory usage.
enerate a multiscale, chunked, multi-dimensional spatial image data structure
that can serialized to OME-NGFF.
Each scale is a scientific Python Xarray spatial-image Dataset, organized
into nodes of an Xarray Datatree.
Installation
pip install multiscale_spatial_image
Usage
import numpy as np
from spatial_image import to_spatial_image
from multiscale_spatial_image import to_multiscale
import zarr
# Image pixels
array = np.random.randint(0, 256, size=(128,128), dtype=np.uint8)
image = to_spatial_image(array)
print(image)
An Xarray spatial-image DataArray. Spatial metadata can also be passed
during construction.
<xarray.SpatialImage 'image' (y: 128, x: 128)>
array([[114, 47, 215, ..., 245, 14, 175],
[ 94, 186, 112, ..., 42, 96, 30],
[133, 170, 193, ..., 176, 47, 8],
...,
[202, 218, 237, ..., 19, 108, 135],
[ 99, 94, 207, ..., 233, 83, 112],
[157, 110, 186, ..., 142, 153, 42]], dtype=uint8)
Coordinates:
* y (y) float64 0.0 1.0 2.0 3.0 4.0 ... 123.0 124.0 125.0 126.0 127.0
* x (x) float64 0.0 1.0 2.0 3.0 4.0 ... 123.0 124.0 125.0 126.0 127.0
# Create multiscale pyramid, downscaling by a factor of 2, then 4
multiscale = to_multiscale(image, [2, 4])
print(multiscale)
A chunked Dask Array MultiscaleSpatialImage Xarray Datatree.
DataTree('multiscales', parent=None)
├── DataTree('scale0')
│ Dimensions: (y: 128, x: 128)
│ Coordinates:
│ * y (y) float64 0.0 1.0 2.0 3.0 4.0 ... 123.0 124.0 125.0 126.0 127.0
│ * x (x) float64 0.0 1.0 2.0 3.0 4.0 ... 123.0 124.0 125.0 126.0 127.0
│ Data variables:
│ image (y, x) uint8 dask.array<chunksize=(128, 128), meta=np.ndarray>
├── DataTree('scale1')
│ Dimensions: (y: 64, x: 64)
│ Coordinates:
│ * y (y) float64 0.5 2.5 4.5 6.5 8.5 ... 118.5 120.5 122.5 124.5 126.5
│ * x (x) float64 0.5 2.5 4.5 6.5 8.5 ... 118.5 120.5 122.5 124.5 126.5
│ Data variables:
│ image (y, x) uint8 dask.array<chunksize=(64, 64), meta=np.ndarray>
└── DataTree('scale2')
Dimensions: (y: 16, x: 16)
Coordinates:
* y (y) float64 3.5 11.5 19.5 27.5 35.5 ... 91.5 99.5 107.5 115.5 123.5
* x (x) float64 3.5 11.5 19.5 27.5 35.5 ... 91.5 99.5 107.5 115.5 123.5
Data variables:
image (y, x) uint8 dask.array<chunksize=(16, 16), meta=np.ndarray>
Store as an Open Microscopy Environment-Next Generation File Format (OME-NGFF)
/ netCDF Zarr store.
It is highly recommended to use dimension_separator='/' in the construction of
the Zarr stores.
store = zarr.storage.DirectoryStore('multiscale.zarr', dimension_separator='/')
multiscale.to_zarr(store)
- A more detailed ITK-based example:

This is a wrapper around a pure C++ core.
Installation
pip install itk-ioomezarrngff
Example
import itk
image = itk.imread('cthead1.png')
itk.imwrite(image, 'ctheaed1.ome.zarr')
in the context of ome-zarr, if you need more sophisticated transforms, you will have to wait a couple of versions I’m afraid — see my RFC-3 1 and the currently stalled but imho close-to-final transformations specification PR.
We will continue to hack on OME-Zarr Coordinate Transformations this weekend – please join us!