I have a series of CT images, now I want to process these images one by one, and save these images processed in memory as a 3D data? Then , I can convert this ITK 3D data to VTK image and visual.
Maybe read your series as a 3D image and use SliceBySliceImageFilter?
Alternatively use a for loop to read and process your images, and use paste filter to assemble them into a 3D volume.
But if you have a 3D image, it is usually desirable to process it as a 3D image rather than a series of 2D slices (that is called 2.5D approach).
OKAY, i Will try! thanks
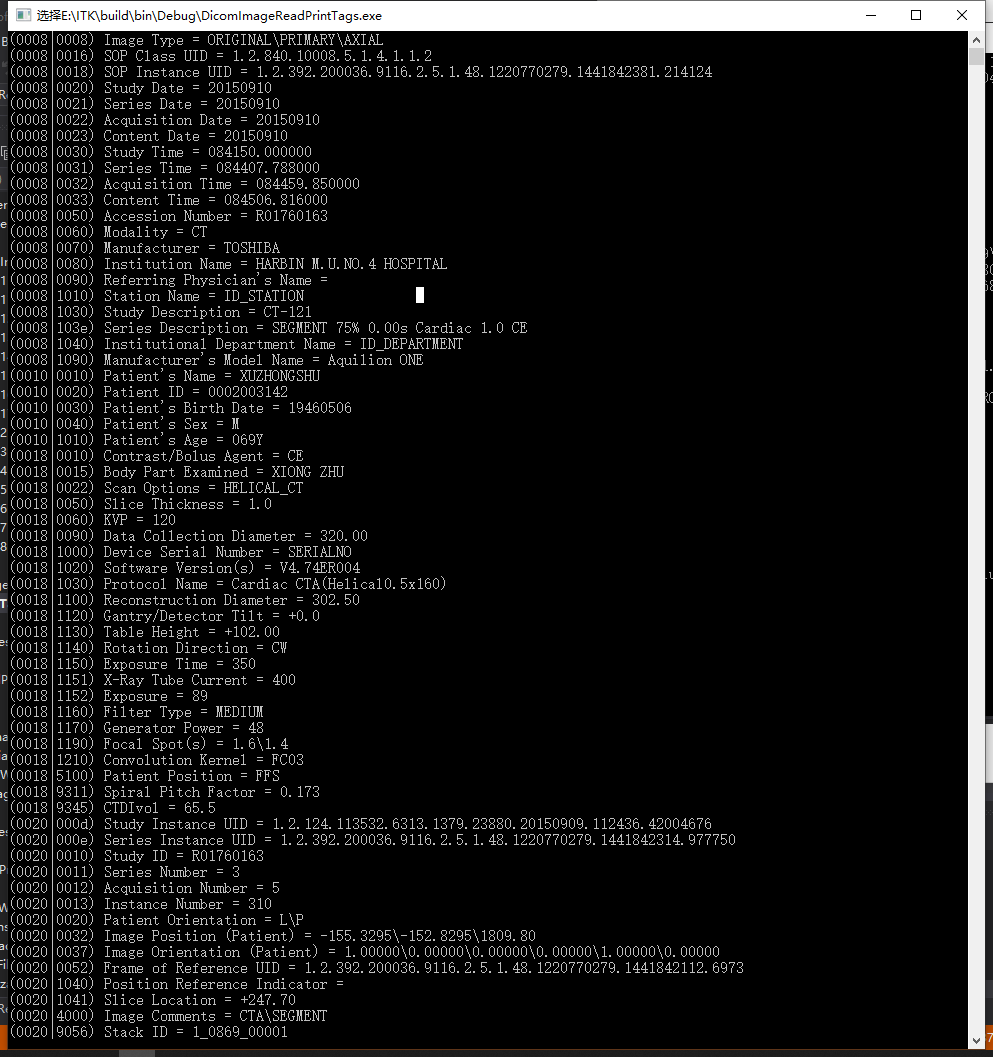
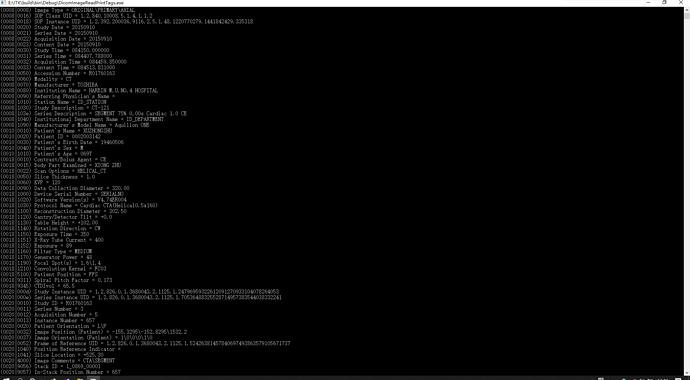
Sorry, I have try the “past filter”,but failed! Then I write processed dicom image as another dicom file,But always 3k Byte losed, and I can’t reconstruct again(use RadiAnt DICOM Viewer ), I thouth the header infomation losed! I also tried the ITK demo “DicomImageReadWrite” and “DicomSeriesReadSeriesWrite”,any suggesitions please!
Hello the code like flow:
#include "itkImageFileReader.h"
#include "itkImageFileWriter.h"
#include "itkGDCMImageIO.h"
#include <list>
#include <fstream>
int main(int argc, char* argv[])
{
typedef signed short InputPixelType;
const unsigned int InputDimension = 2;
typedef itk::Image< InputPixelType, InputDimension > InputImageType;
typedef itk::ImageFileReader< InputImageType > ReaderType;
ReaderType::Pointer reader = ReaderType::New();
reader->SetFileName("C:/input/lung.dcm");
typedef itk::GDCMImageIO ImageIOType;
ImageIOType::Pointer gdcmImageIO = ImageIOType::New();
reader->SetImageIO(gdcmImageIO);
try
{
reader->Update();
}
catch (itk::ExceptionObject & e)
{
std::cerr << "exception in file reader " << std::endl;
std::cerr << e << std::endl;
return EXIT_FAILURE;
}
typedef itk::ImageFileWriter< InputImageType > Writer1Type;
Writer1Type::Pointer writer = Writer1Type::New();
writer->SetFileName("C:/output/lung.dcm");
writer->SetInput(reader->GetOutput());
try
{
writer->Update();
}
catch (itk::ExceptionObject & e)
{
std::cerr << "exception in file writer " << std::endl;
std::cerr << e << std::endl;
return EXIT_FAILURE;
}
return EXIT_SUCCESS;
}
You may need something like writer->SetMetaDataDictionary(reader->GetMetaDataDictionary());. Maybe look at this DicomSeriesReadSeriesWrite example.
Also it is desirable to use backticks for code formatting.
I have run the code ,It is ok, But , I can’t process Image one by one!!! Any suggestions?
I want to process dicom images of one frame of a series.
Hello @wulicheng,
This is actually a good thing. You processed the images and when you save them they should be to a different series, likely with the same study number.
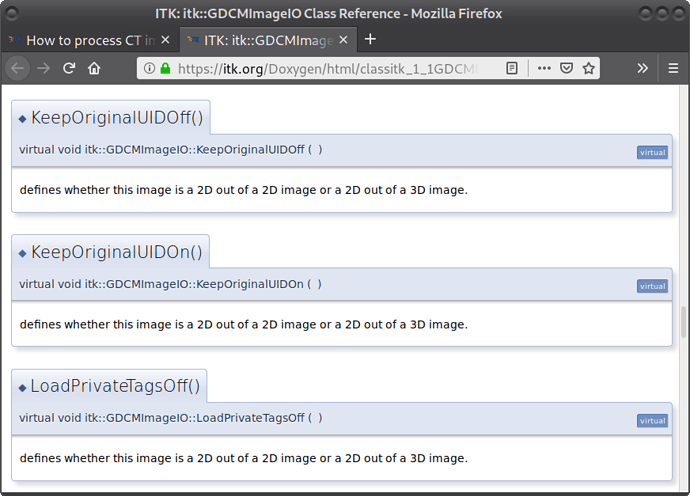
If you still feel you want to retain the study and series numbers you will need to explicitly set the writer to use GDCMImageIO, set the series/study number to what you want in the meta-data dictionary and then configure the GDCMImageIO to use these by calling the KeepOriginalUIDOn method.
thanks very much! I have got it!
Hi @mihail.isakov,
Good catch, didn’t even notice that when pointing to the doxygen.
Looks like something is wrong with the doxygen generation as the documentation in the code is correct.
@dzenanz, any idea what’s wrong with doxygen generation?