Dataset at “https://public.kitware.com/Wiki/TubeTK/Data” have “VascularNetwork.tre” file. How can i open this file using Python?
Hi @Duy_H,
Welcome to ITK! 
To read VascularNetwork.tre,
import itk
file_name = '/tmp/path/to/VascularNetwork.tre'
Dimension = 3
reader = itk.SpatialObjectReader[Dimension].New()
reader.SetFileName(file_name)
reader.Update()
tubes = reader.GetGroup()
print(tubes)
@matt.mccormick thanks for this.
I am trying to convert the .tre into Nifi file. Looking at old TubeTK example files. I tried to use SpatialObjectToImageFilter with not much luck. Any idea how to convert the tre into a binary mask?
Thanks
Ali
@matt.mccormick Thank you so much for the quick response. I am trying to see if I can do it without TubeTK since it’s a real pain to install it. I will see what I can do with this.
Never-less thank you so much for the quick response.
@matt.mccormick I spent a lot of time compiling ITK with Module_TubeTK and I am getting errors.
Targets not yet defined:
ITKBioCell;ITKFEM;itkgdcmopenjp2;ITKIOMesh;ITKReview;IsotropicWavelets
Do you have any suggestions to resolve this? I am following your instructions from this:
Any idea?
Ali
@Stephen_Aylward what are the most up-to-date build instructions for TubeTK?
Build ITK and turn on the TubeTK option.
Only works with ITK master (head) source code.
s
@Stephen_Aylward thanks, but I did that. Compiled ITK, then again for TubeTK, and the following error showed at 95% progress
[ 95%] Generating itkGEAdwImageIOPython.cpp, …/…/…/lib/itkGEAdwImageIOPython.py
swig: …/…/swig/Source/Modules/lang.cxx:1657: virtual int Language::enumDeclaration(Node*): Assertion `Getattr(n, “sym:name”)’ failed.
Aborted (core dumped)
Wrapping/Modules/ITKIOGE/CMakeFiles/ITKIOGEPython.dir/build.make:117: recipe for target ‘Wrapping/Modules/ITKIOGE/itkGEAdwImageIOPython.cpp’ failed
make[2]: *** [Wrapping/Modules/ITKIOGE/itkGEAdwImageIOPython.cpp] Error 134
make[2]: *** Deleting file ‘Wrapping/Modules/ITKIOGE/itkGEAdwImageIOPython.cpp’
CMakeFiles/Makefile2:55091: recipe for target ‘Wrapping/Modules/ITKIOGE/CMakeFiles/ITKIOGEPython.dir/all’ failed
make[1]: *** [Wrapping/Modules/ITKIOGE/CMakeFiles/ITKIOGEPython.dir/all] Error 2
Makefile:162: recipe for target ‘all’ failed
make: *** [all] Error 2
Hi,
Sorry that you’re having troubles. That seems like a bug in the IOGE module.
Are you trying to compile the executables in TubeTK? I’ve got a branch in my private repo to fix some bugs in the apps since the merge, but it does seem like your bug is unrelated. In general, if you don’t need the applications, you should compile ITK with the TubeTK module enabled from the start - there is no need to recompile. The C++ and python interfaces should then work fine.
s
Hi Stephen,
Thanks for following up. To be honest, I just need to convert a series of TRE files into NIFTI mask. I dont even need TubeTK. Do you have any code which uses ITK only to convert TRE files into NIFTI mask?
Thanks
And also, I am getting the above error when I am compiling ITK with TubeTK enabled from start. Based on your suggestion, I thought I have to compile ITK first to build these targets, then recompile. But, again, I dont really need TubeTK, I am only interested to convert a series of TRE file into NIFTI mask.
There is a Jupyter Notebook that converts TRE to an image using only ITK (no TubeTK):
It only generates an image of the centerlines of the tubes, but perhaps that is enough? You could also build upon it to have it generate the radius without too much additional work.
That is surprising that your ITK isn’t compiling! Are you using ITK head/master? Does it compile without TubeTK enabled?
Thanks,
Stephen
Thanks much @Stephen_Aylward
I am trying that example, so far I am getting error:
module ‘itk’ has no attribute ‘CastSpatialObjectFilter’
Regarding the compile, I did try to compile on a fresh server with TubeTK enabled, but it requires those targets which are not available unless I compile the ITK first. Then I can configure the cmake to compile TubeTK and that’s what I did and finally got another error that I mentioned earlier.
Hi,
It seems like you are not compiling ITK master/head, otherwise it would find CastSpatialObjectFilter. If you are using an older version of ITK, then TubeTK is very likely not going to work. Sorry.
Stephen
Stephen,
I am almost certain, but I will double check. Thanks
Ali
@Stephen_Aylward First of all, thanks for all the help.
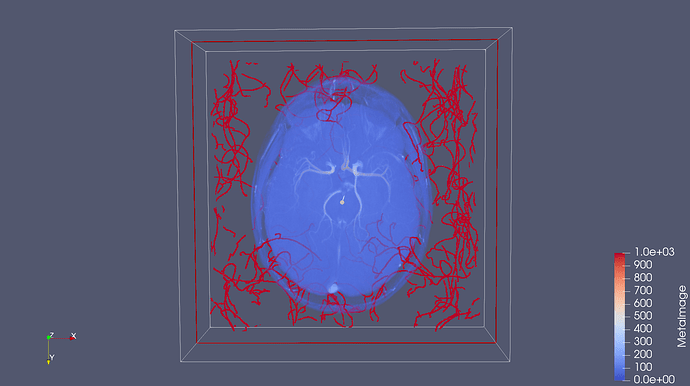
So I managed to convert to nifti, but I am trying to make sense of the centerline position compare to image coordinate. Seems like each object has it’s own coordinate system which is not absolute, is that correct? If so, how compute the world coordinate system for each object? Is there any conversion matrix in each object?
Btw, I am not using cast, I am using the following code:
…
baseSO = sobj[iterChikdID]
baseSOTyped = itkExtras.down_cast(baseSO)
for iterObjectID in range(0,baseSOTyped.GetNumberOfPoints()):
position = baseSOTyped.GetPoint(iterObjectID).GetPositionInObjectSpace()
indx = mra.TransformPhysicalPointToIndex(position)
mra.SetPixel(indx, 100000)
…
Hi
Congratulations on the progress!
Is it correct that you are loading a tre file from one patient and displaying it within the mra from that same patient? Is this one of the mra from the database at kitware.com?
Each patient has a unique coordinate space, so vessels from one patient won’t align with vessels or scans from another patient.
In theory you should be calling GetPositionInWorldSpace() instead of GetPositionInObjectSpace(), but that only matters if you have created a hierarchy of objects. I’d have to see the tre file to determine if that is what is happening.
Hope this helps. Sorry for bevity, typing on a phone while in a forest 
S
@Stephen_Aylward, it wouldnt happened without your help 
I am using the same subject data (Normal002_MRA and it’s tre file). I tried the WorldSpace and getting the following error:
The SpatialObject must be set prior to calling.
I have to figure out how to that. But is there anyway to find the conversion matrix of each object?
I really appreciate your response while in a forest!
Hi
I again suspect that you are seeing an old bug.
When did you clone and update itk last?
S